 |
PDBsum entry 2c75

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
2c75
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.1.4.3.21
- primary-amine oxidase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a primary methyl amine + O2 + H2O = an aldehyde + H2O2 + NH4+
|
 |
 |
 |
 |
 |
primary methyl amine
|
+
|
 O2
O2
|
+
|
H2O
|
=
|
 aldehyde
aldehyde
|
+
|
 H2O2
H2O2
|
+
|
NH4(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.1.4.3.4
- monoamine oxidase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a secondary aliphatic amine + O2 + H2O = a primary amine + an aldehyde + H2O2
|
 |
 |
 |
 |
 |
secondary aliphatic amine
|
+
|
 O2
O2
|
+
|
H2O
|
=
|
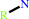 primary amine
primary amine
|
+
|
 aldehyde
aldehyde
|
+
|
 H2O2
H2O2
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
FAD
|
 |
 |
 |
 |
 |
FAD
Bound ligand (Het Group name =
FAD)
corresponds exactly
|
|
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
45:4775-4784
(2006)
|
|
PubMed id:
|
|
|
|

|
| |
|
Functional role of the "aromatic cage" in human monoamine oxidase B: structures and catalytic properties of Tyr435 mutant proteins.
|
|
M.Li,
C.Binda,
A.Mattevi,
D.E.Edmondson.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Current structural results of several flavin-dependent amine oxidizing enzymes
including human monoamine oxidases A and B (MAO A and MAO B) show aromatic amino
acid residues oriented approximately perpendicular to the flavin ring,
suggesting a functional role in catalysis. In the case of human MAO B, two
tyrosyl residues (Y398 and Y435) are found in the substrate binding site on the
re face of the covalent flavin ring [Binda et al. (2002) J. Biol. Chem. 277,
23973-23976]. To probe the functional significance of this structure, Tyr435 in
MAO B was mutated with the amino acids Phe, His, Leu, or Trp, the mutant
proteins expressed in Pichia pastoris, and purified to homogeneity. Each mutant
protein contains covalent FAD and exhibits a high level of catalytic
functionality. No major alterations in active site structures are detected on
comparison of their respective crystal structures with that of WT enzyme. The
relative k(cat)/K(m) values for each mutant enzyme show Y435 > Y435F = Y435L
= Y435H > Y435W. A similar behavior is also observed with the membrane-bound
forms of MAO A and MAO B (MAO A Y444 mutant enzymes are found to be unstable on
membrane extraction). p-Nitrobenzylamine is found to be a poor substrate while
p-nitrophenethylamine is found to be a good substrate for all WT and mutant
forms of MAO B. Analysis of these kinetic and structural data suggests the
function of the "aromatic cage" in MAO to include a steric role in substrate
binding and access to the flavin coenzyme and to increase the nucleophilicity of
the substrate amine moiety. These results are consistent with a proposed polar
nucleophilic mechanism for catalytic amine oxidation.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
G.Kachalova,
K.Decker,
A.Holt,
and
H.D.Bartunik
(2011).
Crystallographic snapshots of the complete reaction cycle of nicotine degradation by an amine oxidase of the monoamine oxidase (MAO) family.
|
| |
Proc Natl Acad Sci U S A,
108,
4800-4805.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
B.K.Arslan,
and
D.E.Edmondson
(2010).
Expression of zebrafish (Danio rerio) monoamine oxidase (MAO) in Pichia pastoris: purification and comparison with human MAO A and MAO B.
|
| |
Protein Expr Purif,
70,
290-297.
|
 |
|
|
|
|
 |
R.V.Dunn,
A.W.Munro,
N.J.Turner,
S.E.Rigby,
and
N.S.Scrutton
(2010).
Tyrosyl radical formation and propagation in flavin dependent monoamine oxidases.
|
| |
Chembiochem,
11,
1228-1231.
|
 |
|
|
|
|
 |
D.E.Edmondson,
C.Binda,
J.Wang,
A.K.Upadhyay,
and
A.Mattevi
(2009).
Molecular and mechanistic properties of the membrane-bound mitochondrial monoamine oxidases.
|
| |
Biochemistry,
48,
4220-4230.
|
 |
|
|
|
|
 |
F.Forneris,
E.Battaglioli,
A.Mattevi,
and
C.Binda
(2009).
New roles of flavoproteins in molecular cell biology: histone demethylase LSD1 and chromatin.
|
| |
FEBS J,
276,
4304-4312.
|
 |
|
|
|
|
 |
J.Wang,
J.Harris,
D.D.Mousseau,
and
D.E.Edmondson
(2009).
Mutagenic probes of the role of Ser209 on the cavity shaping loop of human monoamine oxidase A.
|
| |
FEBS J,
276,
4569-4581.
|
 |
|
|
|
|
 |
A.K.Upadhyay,
and
D.E.Edmondson
(2008).
Characterization of detergent purified recombinant rat liver monoamine oxidase B expressed in Pichia pastoris.
|
| |
Protein Expr Purif,
59,
349-356.
|
 |
|
|
|
|
 |
E.M.Milczek,
D.Bonivento,
C.Binda,
A.Mattevi,
I.A.McDonald,
and
D.E.Edmondson
(2008).
Structural and mechanistic studies of mofegiline inhibition of recombinant human monoamine oxidase B.
|
| |
J Med Chem,
51,
8019-8026.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
E.W.van Hellemond,
M.van Dijk,
D.P.Heuts,
D.B.Janssen,
and
M.W.Fraaije
(2008).
Discovery and characterization of a putrescine oxidase from Rhodococcus erythropolis NCIMB 11540.
|
| |
Appl Microbiol Biotechnol,
78,
455-463.
|
 |
|
|
|
|
 |
K.Ida,
M.Kurabayashi,
M.Suguro,
Y.Hiruma,
T.Hikima,
M.Yamomoto,
and
H.Suzuki
(2008).
Structural basis of proteolytic activation of L-phenylalanine oxidase from Pseudomonas sp. P-501.
|
| |
J Biol Chem,
283,
16584-16590.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.A.Musumeci,
A.K.Arakaki,
D.V.Rial,
D.L.Catalano-Dupuy,
and
E.A.Ceccarelli
(2008).
Modulation of the enzymatic efficiency of ferredoxin-NADP(H) reductase by the amino acid volume around the catalytic site.
|
| |
FEBS J,
275,
1350-1366.
|
 |
|
|
|
|
 |
R.V.Dunn,
K.R.Marshall,
A.W.Munro,
and
N.S.Scrutton
(2008).
The pH dependence of kinetic isotope effects in monoamine oxidase A indicates stabilization of the neutral amine in the enzyme-substrate complex.
|
| |
FEBS J,
275,
3850-3858.
|
 |
|
|
|
|
 |
S.Hruschka,
T.C.Rosen,
S.Yoshida,
K.L.Kirk,
R.Fröhlich,
B.Wibbeling,
and
G.Haufe
(2008).
Fluorinated phenylcyclopropylamines. Part 5: Effects of electron-withdrawing or -donating aryl substituents on the inhibition of monoamine oxidases A and B by 2-aryl-2-fluoro-cyclopropylamines.
|
| |
Bioorg Med Chem,
16,
7148-7166.
|
 |
|
|
|
|
 |
D.E.Edmondson,
C.Binda,
and
A.Mattevi
(2007).
Structural insights into the mechanism of amine oxidation by monoamine oxidases A and B.
|
| |
Arch Biochem Biophys,
464,
269-276.
|
 |
|
|
|
|
 |
K.Yelekçi,
O.Karahan,
and
M.Toprakçi
(2007).
Docking of novel reversible monoamine oxidase-B inhibitors: efficient prediction of ligand binding sites and estimation of inhibitors thermodynamic properties.
|
| |
J Neural Transm,
114,
725-732.
|
 |
|
|
|
|
 |
M.A.Akyüz,
S.S.Erdem,
and
D.E.Edmondson
(2007).
The aromatic cage in the active site of monoamine oxidase B: effect on the structural and electronic properties of bound benzylamine and p-nitrobenzylamine.
|
| |
J Neural Transm,
114,
693-698.
|
 |
|
|
|
|
 |
P.F.Fitzpatrick
(2007).
Insights into the mechanisms of flavoprotein oxidases from kinetic isotope effects.
|
| |
J Labelled Comp Radiopharm,
50,
1016-1025.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

