 |
PDBsum entry 2bv0

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
2bv0
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Crystal structure of protocatechuate 3,4-dioxygenase from acinetobacter sp. Adp1 mutant r133h in complex with protocatechuate.
|
|
Structure:
|
 |
Protocatechuate 3,4-dioxygenase alpha chain. Chain: a. Synonym: protocatechuate 3,4-dioxygenase, 3,4-pcd. Engineered: yes. Mutation: yes. Protocatechuate 3,4-dioxygenase beta chain. Chain: b. Synonym: protocatechuate 3,4-dioxygenase, 3,4-pcd. Engineered: yes
|
|
Source:
|
 |
Acinetobacter calcoaceticus. Organism_taxid: 62977. Strain: adp1. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Biol. unit:
|
 |
 24mer (from PDB file)
24mer (from PDB file)
|
|
Resolution:
|
 |
|
1.80Å
|
R-factor:
|
0.167
|
R-free:
|
0.192
|
|
|
Authors:
|
 |
M.W.Vetting,M.P.Valley,D.A.D'Argenio,L.N.Ornston,J.D.Lipscomb, D.H.Ohlendorf
|
|
Key ref:
|
 |
M.W.Vetting
Crystallgraphic studies of intradiol dioxygenases..
Ph d thesis,
.
|
 |
|
Date:
|
 |
|
20-Jun-05
|
Release date:
|
26-Oct-06
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chains A, B:
E.C.1.13.11.3
- protocatechuate 3,4-dioxygenase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Benzoate Metabolism
|
 |
 |
 |
 |
 |
Reaction:
|
 |
3,4-dihydroxybenzoate + O2 = 3-carboxy-cis,cis-muconate + 2 H+
|
 |
 |
 |
 |
 |
3,4-dihydroxybenzoate
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
 O2
O2
|
=
|
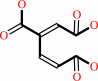 3-carboxy-cis,cis-muconate
3-carboxy-cis,cis-muconate
|
+
|
2
×
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Fe cation
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
|

