 |
PDBsum entry 2acq

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
2acq
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
An anion binding site in human aldose reductase: mechanistic implications for the binding of citrate, cacodylate, and glucose-6- phosphate
|
|
Structure:
|
 |
Aldose reductase. Chain: a. Engineered: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Expressed in: escherichia coli. Expression_system_taxid: 562
|
|
Resolution:
|
 |
|
|
Authors:
|
 |
D.H.Harrison,K.M.Bohren,K.H.Gabbay,G.A.Petsko,D.Ringe
|
Key ref:

|
 |
D.H.Harrison
et al.
(1994).
An anion binding site in human aldose reductase: mechanistic implications for the binding of citrate, cacodylate, and glucose 6-phosphate.
Biochemistry,
33,
2011-2020.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
15-Apr-94
|
Release date:
|
31-Jul-94
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P15121
(ALDR_HUMAN) -
Aldo-keto reductase family 1 member B1 from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
316 a.a.
315 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.1.1.1.21
- aldose reductase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
an alditol + NAD+ = an aldose + NADH + H+
|
|
2.
|
an alditol + NADP+ = an aldose + NADPH + H+
|
|
 |
 |
 |
 |
 |
 alditol
alditol
|
+
|
NAD(+)
Bound ligand (Het Group name = )
matches with 91.67% similarity
|
=
|
 aldose
aldose
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 alditol
alditol
|
+
|
NADP(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
 aldose
aldose
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.1.1.1.300
- NADP-retinol dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
all-trans-retinol + NADP+ = all-trans-retinal + NADPH + H+
|
 |
 |
 |
 |
 |
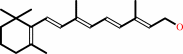 all-trans-retinol
all-trans-retinol
|
+
|
NADP(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
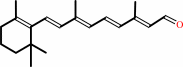 all-trans-retinal
all-trans-retinal
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.1.1.1.372
- D/L-glyceraldehyde reductase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
glycerol + NADP+ = L-glyceraldehyde + NADPH + H+
|
|
2.
|
glycerol + NADP+ = D-glyceraldehyde + NADPH + H+
|
|
 |
 |
 |
 |
 |
 glycerol
glycerol
|
+
|
NADP(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
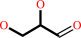 L-glyceraldehyde
L-glyceraldehyde
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 glycerol
glycerol
|
+
|
NADP(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
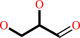 D-glyceraldehyde
D-glyceraldehyde
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 4:
|
 |
E.C.1.1.1.54
- allyl-alcohol dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
allyl alcohol + NADP+ = acrolein + NADPH + H+
|
 |
 |
 |
 |
 |
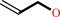 allyl alcohol
allyl alcohol
|
+
|
NADP(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
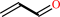 acrolein
acrolein
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
33:2011-2020
(1994)
|
|
PubMed id:
|
|
|
|

|
| |
|
An anion binding site in human aldose reductase: mechanistic implications for the binding of citrate, cacodylate, and glucose 6-phosphate.
|
|
D.H.Harrison,
K.M.Bohren,
D.Ringe,
G.A.Petsko,
K.H.Gabbay.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Aldose reductase is a NADPH-dependent aldo-keto reductase involved in the
pathogenesis of some diabetic and galactosemic complications. The published
crystal structure of human aldose reductase [Wilson et al. (1992) Science 257,
81-84] contains a hitherto unexplained electron density positioned within the
active site pocket facing the nicotinamide ring of the NADPH and other key
active site residues (Tyr48, His110, and Cys298). In this paper we identify the
electron density as citrate, which is present in the crystallization buffer (pH
5.0), and provide confirmatory evidence by both kinetic and crystallographic
experiments. Citrate is an uncompetitive inhibitor in the forward reaction with
respect to aldehyde (reduction of aldehyde), while it is a competitive inhibitor
with respect to alcohol in the backward reaction (oxidation of alcohol),
indicating that it interacts with the enzyme-NADP(+)-product complex. Citrate
can be replaced in the crystalline enzyme complex by cacodylate or glucose
6-phosphate; the structure of each of these complexes shows the specific
molecule bound in the active site. All of the structures have been determined to
a nominal resolution of 1.76 A and refined to R-factors below 18%. While
cacodylate can be bound within the active site under the crystallization
conditions, it does not inhibit the wild-type enzyme in solution. Glucose
6-phosphate, however, is a substrate for aldose reductase. The similar location
of the negative charges of citrate, cacodylate, and glucose 6-phosphate within
the active site suggests an anion-binding site delineated by the C4N of
nicotinamide, the OH of Tyr48, and the N epsilon of His110. The location of
citrate binding in the active site leads to a plausible catalytic mechanism for
aldose reductase.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
G.Bashiri,
C.J.Squire,
N.J.Moreland,
and
E.N.Baker
(2008).
Crystal structures of F420-dependent glucose-6-phosphate dehydrogenase FGD1 involved in the activation of the anti-tuberculosis drug candidate PA-824 reveal the basis of coenzyme and substrate binding.
|
| |
J Biol Chem,
283,
17531-17541.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.G.Olsen,
L.Pedersen,
C.L.Christensen,
O.Olsen,
and
A.Henriksen
(2008).
Barley aldose reductase: structure, cofactor binding, and substrate recognition in the aldo/keto reductase 4C family.
|
| |
Proteins,
71,
1572-1581.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
O.A.Barski,
S.M.Tipparaju,
and
A.Bhatnagar
(2008).
The aldo-keto reductase superfamily and its role in drug metabolism and detoxification.
|
| |
Drug Metab Rev,
40,
553-624.
|
 |
|
|
|
|
 |
Q.Li,
Y.C.Hwang,
R.Ananthakrishnan,
P.J.Oates,
D.Guberski,
and
R.Ramasamy
(2008).
Polyol pathway and modulation of ischemia-reperfusion injury in Type 2 diabetic BBZ rat hearts.
|
| |
Cardiovasc Diabetol,
7,
33.
|
 |
|
|
|
|
 |
M.Biadene,
I.Hazemann,
A.Cousido,
S.Ginell,
A.Joachimiak,
G.M.Sheldrick,
A.Podjarny,
and
T.R.Schneider
(2007).
The atomic resolution structure of human aldose reductase reveals that rearrangement of a bound ligand allows the opening of the safety-belt loop.
|
| |
Acta Crystallogr D Biol Crystallogr,
63,
665-672.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
D.E.Danley
(2006).
Crystallization to obtain protein-ligand complexes for structure-aided drug design.
|
| |
Acta Crystallogr D Biol Crystallogr,
62,
569-575.
|
 |
|
|
|
|
 |
J.M.Brownlee,
E.Carlson,
A.C.Milne,
E.Pape,
and
D.H.Harrison
(2006).
Structural and thermodynamic studies of simple aldose reductase-inhibitor complexes.
|
| |
Bioorg Chem,
34,
424-444.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
E.I.Howard,
R.Sanishvili,
R.E.Cachau,
A.Mitschler,
B.Chevrier,
P.Barth,
V.Lamour,
M.Van Zandt,
E.Sibley,
C.Bon,
D.Moras,
T.R.Schneider,
A.Joachimiak,
and
A.Podjarny
(2004).
Ultrahigh resolution drug design I: details of interactions in human aldose reductase-inhibitor complex at 0.66 A.
|
| |
Proteins,
55,
792-804.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
G.Obmolova,
A.Teplyakov,
P.P.Khil,
A.J.Howard,
R.D.Camerini-Otero,
and
G.L.Gilliland
(2003).
Crystal structure of the Escherichia coli Tas protein, an NADP(H)-dependent aldo-keto reductase.
|
| |
Proteins,
53,
323-325.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
E.Kozma,
E.Brown,
E.M.Ellis,
and
A.J.Lapthorn
(2002).
The crystal structure of rat liver AKR7A1. A dimeric member of the aldo-keto reductase superfamily.
|
| |
J Biol Chem,
277,
16285-16293.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.J.Ondrechen,
J.G.Clifton,
and
D.Ringe
(2001).
THEMATICS: a simple computational predictor of enzyme function from structure.
|
| |
Proc Natl Acad Sci U S A,
98,
12473-12478.
|
 |
|
|
|
|
 |
K.M.Bohren,
and
C.E.Grimshaw
(2000).
The sorbinil trap: a predicted dead-end complex confirms the mechanism of aldose reductase inhibition.
|
| |
Biochemistry,
39,
9967-9974.
|
 |
|
|
|
|
 |
K.V.Ramana,
B.L.Dixit,
S.Srivastava,
G.K.Balendiran,
S.K.Srivastava,
and
A.Bhatnagar
(2000).
Selective recognition of glutathiolated aldehydes by aldose reductase.
|
| |
Biochemistry,
39,
12172-12180.
|
 |
|
|
|
|
 |
Q.Ye,
D.Hyndman,
X.Li,
T.G.Flynn,
and
Z.Jia
(2000).
Crystal structure of CHO reductase, a member of the aldo-keto reductase superfamily.
|
| |
Proteins,
38,
41-48.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.Khurana,
G.Sanli,
D.B.Powers,
S.Anderson,
and
M.Blaber
(2000).
Molecular modeling of substrate binding in wild-type and mutant Corynebacteria 2,5-diketo-D-gluconate reductases.
|
| |
Proteins,
39,
68-75.
|
 |
|
|
|
|
 |
H.Rogniaux,
A.Van Dorsselaer,
P.Barth,
J.F.Biellmann,
J.Barbanton,
M.van Zandt,
B.Chevrier,
E.Howard,
A.Mitschler,
N.Potier,
L.Urzhumtseva,
D.Moras,
and
A.Podjarny
(1999).
Binding of aldose reductase inhibitors: correlation of crystallographic and mass spectrometric studies.
|
| |
J Am Soc Mass Spectrom,
10,
635-647.
|
 |
|
|
|
|
 |
J.M.Gulbis,
S.Mann,
and
R.MacKinnon
(1999).
Structure of a voltage-dependent K+ channel beta subunit.
|
| |
Cell,
97,
943-952.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
L.Costantino,
G.Rastelli,
P.Vianello,
G.Cignarella,
and
D.Barlocco
(1999).
Diabetes complications and their potential prevention: aldose reductase inhibition and other approaches.
|
| |
Med Res Rev,
19,
3.
|
 |
|
|
|
|
 |
L.D.Griffin,
and
S.H.Mellon
(1999).
Selective serotonin reuptake inhibitors directly alter activity of neurosteroidogenic enzymes.
|
| |
Proc Natl Acad Sci U S A,
96,
13512-13517.
|
 |
|
|
|
|
 |
P.J.Oates,
and
B.L.Mylari
(1999).
Aldose reductase inhibitors: therapeutic implications for diabetic complications.
|
| |
Expert Opin Investig Drugs,
8,
2095-2119.
|
 |
|
|
|
|
 |
P.Várnai,
W.G.Richards,
and
P.D.Lyne
(1999).
Modelling the catalytic reaction in human aldose reductase.
|
| |
Proteins,
37,
218-227.
|
 |
|
|
|
|
 |
S.Srivastava,
S.J.Watowich,
J.M.Petrash,
S.K.Srivastava,
and
A.Bhatnagar
(1999).
Structural and kinetic determinants of aldehyde reduction by aldose reductase.
|
| |
Biochemistry,
38,
42-54.
|
 |
|
|
|
|
 |
B.P.Schlegel,
J.M.Jez,
and
T.M.Penning
(1998).
Mutagenesis of 3 alpha-hydroxysteroid dehydrogenase reveals a "push-pull" mechanism for proton transfer in aldo-keto reductases.
|
| |
Biochemistry,
37,
3538-3548.
|
 |
|
|
|
|
 |
J.M.Jez,
and
T.M.Penning
(1998).
Engineering steroid 5 beta-reductase activity into rat liver 3 alpha-hydroxysteroid dehydrogenase.
|
| |
Biochemistry,
37,
9695-9703.
|
 |
|
|
|
|
 |
M.J.Crabbe,
and
D.Goode
(1998).
Aldose reductase: a window to the treatment of diabetic complications?
|
| |
Prog Retin Eye Res,
17,
313-383.
|
 |
|
|
|
|
 |
S.Srivastava,
T.M.Harter,
A.Chandra,
A.Bhatnagar,
S.K.Srivastava,
and
J.M.Petrash
(1998).
Kinetic studies of FR-1, a growth factor-inducible aldo-keto reductase.
|
| |
Biochemistry,
37,
12909-12917.
|
 |
|
|
|
|
 |
Y.S.Lee,
M.Hodoscek,
B.R.Brooks,
and
P.F.Kador
(1998).
Catalytic mechanism of aldose reductase studied by the combined potentials of quantum mechanics and molecular mechanics.
|
| |
Biophys Chem,
70,
203-216.
|
 |
|
|
|
|
 |
A.D.Mesecar,
B.L.Stoddard,
and
D.E.Koshland
(1997).
Orbital steering in the catalytic power of enzymes: small structural changes with large catalytic consequences.
|
| |
Science,
277,
202-206.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.Urzhumtsev,
F.Tête-Favier,
A.Mitschler,
J.Barbanton,
P.Barth,
L.Urzhumtseva,
J.F.Biellmann,
A.Podjarny,
and
D.Moras
(1997).
A 'specificity' pocket inferred from the crystal structures of the complexes of aldose reductase with the pharmaceutically important inhibitors tolrestat and sorbinil.
|
| |
Structure,
5,
601-612.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.J.Bennett,
R.H.Albert,
J.M.Jez,
H.Ma,
T.M.Penning,
and
M.Lewis
(1997).
Steroid recognition and regulation of hormone action: crystal structure of testosterone and NADP+ bound to 3 alpha-hydroxysteroid/dihydrodiol dehydrogenase.
|
| |
Structure,
5,
799-812.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
N.Potier,
P.Barth,
D.Tritsch,
J.F.Biellmann,
and
A.Van Dorsselaer
(1997).
Study of non-covalent enzyme-inhibitor complexes of aldose reductase by electrospray mass spectrometry.
|
| |
Eur J Biochem,
243,
274-282.
|
 |
|
|
|
|
 |
M.J.Bennett,
B.P.Schlegel,
J.M.Jez,
T.M.Penning,
and
M.Lewis
(1996).
Structure of 3 alpha-hydroxysteroid/dihydrodiol dehydrogenase complexed with NADP+.
|
| |
Biochemistry,
35,
10702-10711.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.J.Crabbe,
D.Goode,
and
S.P.Wolff
(1996).
Aldose reductase: black sheep or skeleton in the cupboard?
|
| |
Nat Struct Biol,
3,
659-661.
|
 |
|
|
|
|
 |
M.von Itzstein,
and
P.Colman
(1996).
Design and synthesis of carbohydrate-based inhibitors of protein-carbohydrate interactions.
|
| |
Curr Opin Struct Biol,
6,
703-709.
|
 |
|
|
|
|
 |
N.Tanaka,
T.Nonaka,
T.Tanabe,
T.Yoshimoto,
D.Tsuru,
and
Y.Mitsui
(1996).
Crystal structures of the binary and ternary complexes of 7 alpha-hydroxysteroid dehydrogenase from Escherichia coli.
|
| |
Biochemistry,
35,
7715-7730.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
O.A.Barski,
K.H.Gabbay,
and
K.M.Bohren
(1996).
The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity.
|
| |
Biochemistry,
35,
14276-14280.
|
 |
|
|
|
|
 |
S.Janecek
(1996).
Invariant glycines and prolines flanking in loops the strand beta 2 of various (alpha/beta)8-barrel enzymes: a hidden homology?
|
| |
Protein Sci,
5,
1136-1143.
|
 |
|
|
|
|
 |
T.Nakano,
and
J.M.Petrash
(1996).
Kinetic and spectroscopic evidence for active site inhibition of human aldose reductase.
|
| |
Biochemistry,
35,
11196-11202.
|
 |
|
|
|
|
 |
D.A.Carper,
T.C.Hohman,
and
S.E.Old
(1995).
Residues affecting the catalysis and inhibition of rat lens aldose reductase.
|
| |
Biochim Biophys Acta,
1246,
67-73.
|
 |
|
|
|
|
 |
D.H.Harrison
(1995).
All in the family.
|
| |
Nat Struct Biol,
2,
719-720.
|
 |
|
|
|
|
 |
O.el-Kabbani,
K.Judge,
S.L.Ginell,
D.A.Myles,
L.J.DeLucas,
and
T.G.Flynn
(1995).
Structure of porcine aldehyde reductase holoenzyme.
|
| |
Nat Struct Biol,
2,
687-692.
|
 |
|
|
|
|
 |
T.J.Kubiseski,
and
T.G.Flynn
(1995).
Studies on human aldose reductase. Probing the role of arginine 268 by site-directed mutagenesis.
|
| |
J Biol Chem,
270,
16911-16917.
|
 |
|
|
|
|
 |
V.J.Demopoulos,
and
E.Rekka
(1995).
Isomeric benzoylpyrroleacetic acids: some structural aspects for aldose reductase inhibitory and anti-inflammatory activities.
|
| |
J Pharm Sci,
84,
79-82.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

