 |
PDBsum entry 2ac4

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Lyase
|
 |
|
Title:
|
 |
Crystal structure of the his183cys mutant variant of bacillus subtilis ferrochelatase
|
|
Structure:
|
 |
Ferrochelatase. Chain: a. Synonym: protoheme ferro-lyase, heme synthetase. Engineered: yes. Mutation: yes
|
|
Source:
|
 |
Bacillus subtilis. Organism_taxid: 1423. Gene: hemh, hemf. Expressed in: escherichia coli. Expression_system_taxid: 562
|
|
Resolution:
|
 |
|
2.10Å
|
R-factor:
|
0.214
|
R-free:
|
0.276
|
|
|
Authors:
|
 |
S.Shipovskov,T.Karlberg,M.Fodje,M.D.Hansson,G.C.Ferreira,M.Hansson, C.T.Reimann,S.Al-Karadaghi
|
Key ref:

|
 |
S.Shipovskov
et al.
(2005).
Metallation of the transition-state inhibitor N-methyl mesoporphyrin by ferrochelatase: implications for the catalytic reaction mechanism.
J Mol Biol,
352,
1081-1090.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
18-Jul-05
|
Release date:
|
20-Sep-05
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P32396
(HEMH_BACSU) -
Coproporphyrin III ferrochelatase from Bacillus subtilis (strain 168)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
310 a.a.
309 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.99.1.9
- coproporphyrin ferrochelatase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
Fe-coproporphyrin III + 2 H+ = coproporphyrin III + Fe2+
|
 |
 |
 |
 |
 |
Fe-coproporphyrin III
|
+
|
2
×
H(+)
|
=
|
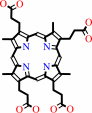 coproporphyrin III
coproporphyrin III
|
+
|
Fe(2+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
352:1081-1090
(2005)
|
|
PubMed id:
|
|
|
|

|
| |
|
Metallation of the transition-state inhibitor N-methyl mesoporphyrin by ferrochelatase: implications for the catalytic reaction mechanism.
|
|
S.Shipovskov,
T.Karlberg,
M.Fodje,
M.D.Hansson,
G.C.Ferreira,
M.Hansson,
C.T.Reimann,
S.Al-Karadaghi.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Insertion of metals into various tetrapyrroles is catalysed by a group of
enzymes called chelatases, e.g. nickel, cobalt, magnesium and ferro-chelatase.
It has been proposed that catalytic metallation includes distorting the
porphyrin substrate by the enzyme towards a transition state-like geometry in
which at least one of the pyrrole rings will be available for metal chelation.
Here, we present a study of metal insertion into the transition-state inhibitor
of protoporphyrin IX ferrochelatase, N-methyl mesoporphyrin (N-MeMP), by
time-resolved crystallography and mass spectrometry with and without the
presence of ferrochelatase. The results show that metallation of N-MeMP has a
very limited effect on the conformation of the residues that participate in
porphyrin and metal binding. These findings support theoretical data, which
indicate that product release is controlled largely by the strain created by
metal insertion into the distorted porphyrin. The results suggest that, similar
to non-catalytic metallation of N-MeMP, the ferrochelatase-assisted metallation
depends on the ligand exchange rate for the respective metal. Moreover,
ferrochelatase catalyses insertion of Cu(II) and Zn(II) into N-MeMP with a rate
that is about 20 times faster than non-enzymatic metallation in solution,
suggesting that the catalytic strategy of ferrochelatase includes a stage of
acceleration of the rate of ligand exchange for the metal substrate. The greater
efficiency of N-MeMP metallation by Cu(II), as compared to Zn(II), contrasts
with the K(m) values for Zn(II) (17 microM) and Cu(II) (170 microM) obtained for
metallation of protoporphyrin IX. We suggest that this difference in metal
specificity depends on the type of distortion imposed by the enzyme on
protoporphyrin IX, which is different from the intrinsic non-planar distortion
of N-MeMP. A mechanism of control of metal specificity by porphyrin distortion
may be general for different chelatases, and may have common features with the
mechanism of metal specificity in crown ethers.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 3.
Figure 3. Isotopic patterns of N-MeMP and its metallated
forms. Theoretical (top row) and experimentally observed (bottom
row) isotopic patterns for (a) N-MeMP, (b) N-MeMP:Cu(II) and (c)
MP:Cu(II).
|
 |
Figure 5.
Figure 5. (a) Metal insertion into N-MeMP in the presence
and in the absence of enzyme. The relative intensity of mass
spectral signals showing Cu(II) insertion into N-MeMP in
solution in the absence (circles) and in the presence (squares)
of B. subtilis ferrochelatase is shown. Zn(II) insertion in the
presence of ferrochelatase is shown as triangles. (b) Cu(II)
insertion into N-MeMP in the presence of B. subtilis
ferrochelatase mutants H183C (diamonds) and Y13F (triangles),
compared to metallation by the wild-type enzyme (squares).
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(2005,
352,
1081-1090)
copyright 2005.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
M.D.Hansson,
T.Karlberg,
C.A.Söderberg,
S.Rajan,
M.J.Warren,
S.Al-Karadaghi,
S.E.Rigby,
and
M.Hansson
(2011).
Bacterial ferrochelatase turns human: Tyr13 determines the apparent metal specificity of Bacillus subtilis ferrochelatase.
|
| |
J Biol Inorg Chem,
16,
235-242.
|
 |
|
|
|
|
 |
N.R.McIntyre,
R.Franco,
J.A.Shelnutt,
and
G.C.Ferreira
(2011).
Nickel(II) chelatase variants directly evolved from murine ferrochelatase: porphyrin distortion and kinetic mechanism.
|
| |
Biochemistry,
50,
1535-1544.
|
 |
|
|
|
|
 |
T.T.Chau,
M.Ishigaki,
T.Kataoka,
and
S.Taketani
(2010).
Porcine ferrochelatase: the relationship between iron-removal reaction and the conversion of heme to Zn-protoporphyrin.
|
| |
Biosci Biotechnol Biochem,
74,
1415-1420.
|
 |
|
|
|
|
 |
T.Karlberg,
M.D.Hansson,
R.K.Yengo,
R.Johansson,
H.O.Thorvaldsen,
G.C.Ferreira,
M.Hansson,
and
S.Al-Karadaghi
(2008).
Porphyrin binding and distortion and substrate specificity in the ferrochelatase reaction: the role of active site residues.
|
| |
J Mol Biol,
378,
1074-1083.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.E.Medlock,
T.A.Dailey,
T.A.Ross,
H.A.Dailey,
and
W.N.Lanzilotta
(2007).
A pi-helix switch selective for porphyrin deprotonation and product release in human ferrochelatase.
|
| |
J Mol Biol,
373,
1006-1016.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Shipovskov,
and
C.T.Reimann
(2007).
Electrospray ionization mass spectrometry in enzymology: uncovering the mechanisms of two-substrate reactions.
|
| |
Analyst,
132,
397-402.
|
 |
|
|
|
|
 |
S.Al-Karadaghi,
R.Franco,
M.Hansson,
J.A.Shelnutt,
G.Isaya,
and
G.C.Ferreira
(2006).
Chelatases: distort to select?
|
| |
Trends Biochem Sci,
31,
135-142.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

