 |
PDBsum entry 2z4y

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Transferase/transferase inhibitor
|
PDB id
|
|

|

|
2z4y
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase/transferase inhibitor
|
 |
|
Title:
|
 |
S. Cerevisiae geranylgeranyl pyrophosphate synthase in complex with magnesium and bph-252
|
|
Structure:
|
 |
Geranylgeranyl pyrophosphate synthetase. Chain: a, b. Synonym: ggpp synthetase, ggppsase, geranylgeranyl diphosphate synthase, bet2 suppressor protein 1. Ec: 2.5.1.30, 2.5.1.1, 2.5.1.10, 2.5.1.29. Engineered: yes
|
|
Source:
|
 |
Saccharomyces cerevisiae. Baker's yeast. Organism_taxid: 4932. Expressed in: escherichia coli bl21(de3). Expression_system_taxid: 469008.
|
|
Resolution:
|
 |
|
2.10Å
|
R-factor:
|
0.181
|
R-free:
|
0.232
|
|
|
Authors:
|
 |
C.K.-M.Chen,R.T.Guo,M.Hudock,R.Cao,E.Oldfield,A.H.-J.Wang
|
|
Key ref:
|
 |
C.K-M Chen
et al.
(2008).
Inhibition of geranylgeranyl diphosphate synthase by bisphosphonates: a crystallographic and computational investigation.
J Med Chem,
51,
5594-5607.
PubMed id:
|
 |
|
Date:
|
 |
|
26-Jun-07
|
Release date:
|
01-Jul-08
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q12051
(GGPPS_YEAST) -
Geranylgeranyl pyrophosphate synthase BTS1 from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
335 a.a.
291 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.2.5.1.1
- dimethylallyltranstransferase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Terpenoid biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
isopentenyl diphosphate + dimethylallyl diphosphate = (2E)- geranyl diphosphate + diphosphate
|
 |
 |
 |
 |
 |
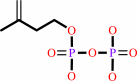 isopentenyl diphosphate
isopentenyl diphosphate
|
+
|
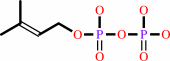 dimethylallyl diphosphate
dimethylallyl diphosphate
|
=
|
(2E)- geranyl diphosphate
|
+
|
 diphosphate
diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.2.5.1.10
- (2E,6E)-farnesyl diphosphate synthase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
|
 |
 |
 |
 |
 |
Reaction:
|
 |
isopentenyl diphosphate + (2E)-geranyl diphosphate = (2E,6E)-farnesyl diphosphate + diphosphate
|
 |
 |
 |
 |
 |
 isopentenyl diphosphate
isopentenyl diphosphate
|
+
|
(2E)-geranyl diphosphate
|
=
|
 (2E,6E)-farnesyl diphosphate
(2E,6E)-farnesyl diphosphate
|
+
|
 diphosphate
diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.2.5.1.29
- geranylgeranyl diphosphate synthase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
|
 |
 |
 |
 |
 |
Reaction:
|
 |
isopentenyl diphosphate + (2E,6E)-farnesyl diphosphate = (2E,6E,10E)- geranylgeranyl diphosphate + diphosphate
|
 |
 |
 |
 |
 |
 isopentenyl diphosphate
isopentenyl diphosphate
|
+
|
 (2E,6E)-farnesyl diphosphate
(2E,6E)-farnesyl diphosphate
|
=
|
(2E,6E,10E)- geranylgeranyl diphosphate
|
+
|
 diphosphate
diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
J Med Chem
51:5594-5607
(2008)
|
|
PubMed id:
|
|
|
|

|
| |
|
Inhibition of geranylgeranyl diphosphate synthase by bisphosphonates: a crystallographic and computational investigation.
|
|
C.K-M Chen,
M.P.Hudock,
Y.Zhang,
R.T.Guo,
R.Cao,
J.H.No,
P.H.Liang,
T.P.Ko,
T.H.Chang,
S.C.Chang,
Y.Song,
J.Axelson,
A.Kumar,
A.H.Wang,
E.Oldfield.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
We report the X-ray structures of several bisphosphonate inhibitors of
geranylgeranyl diphosphate synthase, a target for anticancer drugs.
Bisphosphonates containing unbranched side chains bind to either the farnesyl
diphosphate (FPP) substrate site, the geranylgeranyl diphosphate (GGPP) product
site, and in one case, both sites, with the bisphosphonate moiety interacting
with 3 Mg (2+) that occupy the same position as found in FPP synthase. However,
each of three "V-shaped" bisphosphonates bind to both the FPP and GGPP sites.
Using the Glide program, we reproduced the binding modes of 10 bisphosphonates
with an rms error of 1.3 A. Activities of the bisphosphonates in GGPPS
inhibition were predicted with an overall error of 2x by using a comparative
molecular similarity analysis based on a docked-structure alignment. These
results show that some GGPPS inhibitors can occupy both substrate and product
site and that binding modes as well as activity can be accurately predicted,
facilitating the further development of GGPPS inhibitors as anticancer agents.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
J.D.Artz,
A.K.Wernimont,
J.E.Dunford,
M.Schapira,
A.Dong,
Y.Zhao,
J.Lew,
R.G.Russell,
F.H.Ebetino,
U.Oppermann,
and
R.Hui
(2011).
Molecular characterization of a novel geranylgeranyl pyrophosphate synthase from Plasmodium parasites.
|
| |
J Biol Chem,
286,
3315-3322.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.P.Singh,
Y.Zhang,
J.H.No,
R.Docampo,
V.Nussenzweig,
and
E.Oldfield
(2010).
Lipophilic bisphosphonates are potent inhibitors of Plasmodium liver-stage growth.
|
| |
Antimicrob Agents Chemother,
54,
2987-2993.
|
 |
|
|
|
|
 |
E.Oldfield
(2010).
Targeting isoprenoid biosynthesis for drug discovery: bench to bedside.
|
| |
Acc Chem Res,
43,
1216-1226.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

