 |
PDBsum entry 2uui

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Lyase
|
 |
|
Title:
|
 |
Crystal structure of human leukotriene c4 synthase
|
|
Structure:
|
 |
Leukotriene c4 synthase. Chain: a. Fragment: residues 2-150. Synonym: leukotrienE-C(4) synthase, ltc4 synthase. Engineered: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Expressed in: pichia pastoris. Expression_system_taxid: 4922.
|
|
Resolution:
|
 |
|
2.00Å
|
R-factor:
|
0.198
|
R-free:
|
0.237
|
|
|
Authors:
|
 |
D.Martinez Molina,A.Wetterholm,A.Kohl,A.A.Mccarthy,D.Niegowski, E.Ohlson,T.Hammarberg,S.Eshaghi,J.Z.Haeggstrom,P.Nordlund
|
Key ref:

|
 |
D.Martinez Molina
et al.
(2007).
Structural basis for synthesis of inflammatory mediators by human leukotriene C4 synthase.
Nature,
448,
613-616.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
02-Mar-07
|
Release date:
|
17-Jul-07
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q16873
(LTC4S_HUMAN) -
Leukotriene C4 synthase from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
150 a.a.
156 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.2.5.1.-
- ?????
|
|
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.4.4.1.20
- leukotriene-C4 synthase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
|
 |
 |
 |
 |
 |
Reaction:
|
 |
leukotriene C4 = leukotriene A4 + glutathione
|
 |
 |
 |
 |
 |
leukotriene C4
Bound ligand (Het Group name = )
matches with 41.86% similarity
|
=
|
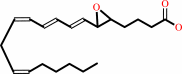 leukotriene A4
leukotriene A4
|
+
|
glutathione
Bound ligand (Het Group name = )
matches with 48.72% similarity
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Nature
448:613-616
(2007)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural basis for synthesis of inflammatory mediators by human leukotriene C4 synthase.
|
|
D.Martinez Molina,
A.Wetterholm,
A.Kohl,
A.A.McCarthy,
D.Niegowski,
E.Ohlson,
T.Hammarberg,
S.Eshaghi,
J.Z.Haeggström,
P.Nordlund.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Cysteinyl leukotrienes are key mediators in inflammation and have an important
role in acute and chronic inflammatory diseases of the cardiovascular and
respiratory systems, in particular bronchial asthma. In the biosynthesis of
cysteinyl leukotrienes, conversion of arachidonic acid forms the unstable
epoxide leukotriene A4 (LTA4). This intermediate is conjugated with glutathione
(GSH) to produce leukotriene C4 (LTC4) in a reaction catalysed by LTC4 synthase:
this reaction is the key step in cysteinyl leukotriene formation. Here we
present the crystal structure of the human LTC4 synthase in its apo and
GSH-complexed forms to 2.00 and 2.15 A resolution, respectively. The structure
reveals a homotrimer, where each monomer is composed of four transmembrane
segments. The structure of the enzyme in complex with substrate reveals that the
active site enforces a horseshoe-shaped conformation on GSH, and effectively
positions the thiol group for activation by a nearby arginine at the
membrane-enzyme interface. In addition, the structure provides a model for how
the omega-end of the lipophilic co-substrate is pinned at one end of a
hydrophobic cleft, providing a molecular 'ruler' to align the reactive epoxide
at the thiol of glutathione. This provides new structural insights into the
mechanism of LTC4 formation, and also suggests that the observed binding and
activation of GSH might be common for a family of homologous proteins important
for inflammatory and detoxification responses.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1: LTC[4] synthase overall structure. a, Left, front
view of LTC[4] synthase protomer in ribbon representation
showing helices 1–5 in successive colouring. The entire LTC[4]
synthase polypeptide, except for the last residue (the last four
in the GSH-complexed structure), can be traced in the electron
density maps. Right, the homotrimer with protomers coloured
orange, green and blue. Bound glutathione, indicating the
position of the active site, is in ball and stick
representation. Approximate membrane positions indicated by blue
lines. b, Cytosolic view of the trimer, showing the position of
the three active sites with loop 1 covering each binding pocket.
Glutathione is shown in ball and stick representation together
with modelled detergent (DDM).
|
 |
Figure 3.
Figure 3: Glutathione binding. a, Electron density map in
blue (F[o] - F[c], contoured at 3  and
phased with the apo structure before refinement) for bound
glutathione shown in ball and stick representation. Interacting
side chains are coloured by monomer and labelled accordingly.
Chemical bonds to glutathione are drawn as dashed lines. A
coordinated water molecule is shown as a light blue sphere. b,
Superposition of the active sites in the apo and
glutathione-bound structures in blue and light brown,
respectively. Glutathione is shown in orange stick
representation and the sulphate molecule from the apo structure
in yellow. and
phased with the apo structure before refinement) for bound
glutathione shown in ball and stick representation. Interacting
side chains are coloured by monomer and labelled accordingly.
Chemical bonds to glutathione are drawn as dashed lines. A
coordinated water molecule is shown as a light blue sphere. b,
Superposition of the active sites in the apo and
glutathione-bound structures in blue and light brown,
respectively. Glutathione is shown in orange stick
representation and the sulphate molecule from the apo structure
in yellow.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Macmillan Publishers Ltd:
Nature
(2007,
448,
613-616)
copyright 2007.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.Oakley
(2011).
Glutathione transferases: a structural perspective.
|
| |
Drug Metab Rev,
43,
138-151.
|
 |
|
|
|
|
 |
L.G.Higgins,
and
J.D.Hayes
(2011).
Mechanisms of induction of cytosolic and microsomal glutathione transferase (GST) genes by xenobiotics and pro-inflammatory agents.
|
| |
Drug Metab Rev,
43,
92.
|
 |
|
|
|
|
 |
N.C.Gilbert,
S.G.Bartlett,
M.T.Waight,
D.B.Neau,
W.E.Boeglin,
A.R.Brash,
and
M.E.Newcomer
(2011).
The structure of human 5-lipoxygenase.
|
| |
Science,
331,
217-219.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
R.Morgenstern,
J.Zhang,
and
K.Johansson
(2011).
Microsomal glutathione transferase 1: mechanism and functional roles.
|
| |
Drug Metab Rev,
43,
300-306.
|
 |
|
|
|
|
 |
Y.Fan,
L.Shi,
V.Ladizhansky,
and
L.S.Brown
(2011).
Uniform isotope labeling of a eukaryotic seven-transmembrane helical protein in yeast enables high-resolution solid-state NMR studies in the lipid environment.
|
| |
J Biomol NMR,
49,
151-161.
|
 |
|
|
|
|
 |
A.Rinaldo-Matthis,
A.Wetterholm,
D.Martinez Molina,
J.Holm,
D.Niegowski,
E.Ohlson,
P.Nordlund,
R.Morgenstern,
and
J.Z.Haeggström
(2010).
Arginine 104 is a key catalytic residue in leukotriene C4 synthase.
|
| |
J Biol Chem,
285,
40771-40776.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
G.Zhao,
M.C.Johnson,
J.R.Schnell,
Y.Kanaoka,
W.Haase,
D.Irikura,
B.K.Lam,
and
I.Schmidt-Krey
(2010).
Two-dimensional crystallization conditions of human leukotriene C4 synthase requiring adjustment of a particularly large combination of specific parameters.
|
| |
J Struct Biol,
169,
450-454.
|
 |
|
|
|
|
 |
K.McLuskey,
A.W.Roszak,
Y.Zhu,
and
N.W.Isaacs
(2010).
Crystal structures of all-alpha type membrane proteins.
|
| |
Eur Biophys J,
39,
723-755.
|
 |
|
|
|
|
 |
K.R.Vinothkumar,
and
R.Henderson
(2010).
Structures of membrane proteins.
|
| |
Q Rev Biophys,
43,
65.
|
 |
|
|
|
|
 |
M.Miyano,
H.Ago,
H.Saino,
T.Hori,
and
K.Ida
(2010).
Internally bridging water molecule in transmembrane alpha-helical kink.
|
| |
Curr Opin Struct Biol,
20,
456-463.
|
 |
|
|
|
|
 |
S.C.Pawelzik,
N.R.Uda,
L.Spahiu,
C.Jegerschöld,
P.Stenberg,
H.Hebert,
R.Morgenstern,
and
P.J.Jakobsson
(2010).
Identification of key residues determining species differences in inhibitor binding of microsomal prostaglandin E synthase-1.
|
| |
J Biol Chem,
285,
29254-29261.
|
 |
|
|
|
|
 |
T.S.Hallstrand,
and
W.R.Henderson
(2010).
An update on the role of leukotrienes in asthma.
|
| |
Curr Opin Allergy Clin Immunol,
10,
60-66.
|
 |
|
|
|
|
 |
A.Maekawa,
B.Balestrieri,
K.F.Austen,
and
Y.Kanaoka
(2009).
GPR17 is a negative regulator of the cysteinyl leukotriene 1 receptor response to leukotriene D4.
|
| |
Proc Natl Acad Sci U S A,
106,
11685-11690.
|
 |
|
|
|
|
 |
J.Alander,
J.Lengqvist,
P.J.Holm,
R.Svensson,
P.Gerbaux,
R.H.Heuvel,
H.Hebert,
W.J.Griffiths,
R.N.Armstrong,
and
R.Morgenstern
(2009).
Microsomal glutathione transferase 1 exhibits one-third-of-the-sites-reactivity towards glutathione.
|
| |
Arch Biochem Biophys,
487,
42-48.
|
 |
|
|
|
|
 |
L.Xing,
R.G.Kurumbail,
R.B.Frazier,
M.S.Davies,
H.Fujiwara,
R.A.Weinberg,
J.K.Gierse,
N.Caspers,
J.S.Carter,
J.J.McDonald,
W.M.Moore,
and
M.L.Vazquez
(2009).
Homo-timeric structural model of human microsomal prostaglandin E synthase-1 and characterization of its substrate/inhibitor binding interactions.
|
| |
J Comput Aided Mol Des,
23,
13-24.
|
 |
|
|
|
|
 |
M.Freigassner,
H.Pichler,
and
A.Glieder
(2009).
wTuning microbial hosts for membrane protein production.
|
| |
Microb Cell Fact,
8,
69.
|
 |
|
|
|
|
 |
M.W.Buczynski,
D.S.Dumlao,
and
E.A.Dennis
(2009).
Thematic Review Series: Proteomics. An integrated omics analysis of eicosanoid biology.
|
| |
J Lipid Res,
50,
1015-1038.
|
 |
|
|
|
|
 |
N.Bonander,
R.A.Darby,
L.Grgic,
N.Bora,
J.Wen,
S.Brogna,
D.R.Poyner,
M.A.O'Neill,
and
R.M.Bill
(2009).
Altering the ribosomal subunit ratio in yeast maximizes recombinant protein yield.
|
| |
Microb Cell Fact,
8,
10.
|
 |
|
|
|
|
 |
T.Hammarberg,
M.Hamberg,
A.Wetterholm,
H.Hansson,
B.Samuelsson,
and
J.Z.Haeggström
(2009).
Mutation of a Critical Arginine in Microsomal Prostaglandin E Synthase-1 Shifts the Isomerase Activity to a Reductase Activity That Converts Prostaglandin H2 into Prostaglandin F2{alpha}.
|
| |
J Biol Chem,
284,
301-305.
|
 |
|
|
|
|
 |
T.Shimizu
(2009).
Lipid mediators in health and disease: enzymes and receptors as therapeutic targets for the regulation of immunity and inflammation.
|
| |
Annu Rev Pharmacol Toxicol,
49,
123-150.
|
 |
|
|
|
|
 |
W.D.Van Horn,
H.J.Kim,
C.D.Ellis,
A.Hadziselimovic,
E.S.Sulistijo,
M.D.Karra,
C.Tian,
F.D.Sönnichsen,
and
C.R.Sanders
(2009).
Solution nuclear magnetic resonance structure of membrane-integral diacylglycerol kinase.
|
| |
Science,
324,
1726-1729.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.K.Mandal,
P.B.Jones,
A.M.Bair,
P.Christmas,
D.Miller,
T.T.Yamin,
D.Wisniewski,
J.Menke,
J.F.Evans,
B.T.Hyman,
B.Bacskai,
M.Chen,
D.M.Lee,
B.Nikolic,
and
R.J.Soberman
(2008).
The nuclear membrane organization of leukotriene synthesis.
|
| |
Proc Natl Acad Sci U S A,
105,
20434-20439.
|
 |
|
|
|
|
 |
A.Maekawa,
Y.Kanaoka,
W.Xing,
and
K.F.Austen
(2008).
Functional recognition of a distinct receptor preferential for leukotriene E4 in mice lacking the cysteinyl leukotriene 1 and 2 receptors.
|
| |
Proc Natl Acad Sci U S A,
105,
16695-16700.
|
 |
|
|
|
|
 |
C.Jegerschöld,
S.C.Pawelzik,
P.Purhonen,
P.Bhakat,
K.R.Gheorghe,
N.Gyobu,
K.Mitsuoka,
R.Morgenstern,
P.J.Jakobsson,
and
H.Hebert
(2008).
Structural basis for induced formation of the inflammatory mediator prostaglandin E2.
|
| |
Proc Natl Acad Sci U S A,
105,
11110-11115.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
D.J.Müller,
N.Wu,
and
K.Palczewski
(2008).
Vertebrate membrane proteins: structure, function, and insights from biophysical approaches.
|
| |
Pharmacol Rev,
60,
43-78.
|
 |
|
|
|
|
 |
D.Veesler,
S.Blangy,
C.Cambillau,
and
G.Sciara
(2008).
There is a baby in the bath water: AcrB contamination is a major problem in membrane-protein crystallization.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
64,
880-885.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
E.P.Carpenter,
K.Beis,
A.D.Cameron,
and
S.Iwata
(2008).
Overcoming the challenges of membrane protein crystallography.
|
| |
Curr Opin Struct Biol,
18,
581-586.
|
 |
|
|
|
|
 |
F.Forneris,
and
A.Mattevi
(2008).
Enzymes without borders: mobilizing substrates, delivering products.
|
| |
Science,
321,
213-216.
|
 |
|
|
|
|
 |
G.Langer,
S.X.Cohen,
V.S.Lamzin,
and
A.Perrakis
(2008).
Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7.
|
| |
Nat Protoc,
3,
1171-1179.
|
 |
|
|
|
|
 |
K.F.Ma,
H.Y.Yang,
Z.Chen,
L.Y.Qi,
D.Y.Zhu,
and
Y.J.Lou
(2008).
Enhanced expressions and activations of leukotriene C4 synthesis enzymes in D-galactosamine/lipopolysaccharide-induced rat fulminant hepatic failure model.
|
| |
World J Gastroenterol,
14,
2748-2756.
|
 |
|
|
|
|
 |
M.Jamshad,
S.Rajesh,
Z.Stamataki,
J.A.McKeating,
T.Dafforn,
M.Overduin,
and
R.M.Bill
(2008).
Structural characterization of recombinant human CD81 produced in Pichia pastoris.
|
| |
Protein Expr Purif,
57,
206-216.
|
 |
|
|
|
|
 |
O.Haworth,
M.Cernadas,
R.Yang,
C.N.Serhan,
and
B.D.Levy
(2008).
Resolvin E1 regulates interleukin 23, interferon-gamma and lipoxin A4 to promote the resolution of allergic airway inflammation.
|
| |
Nat Immunol,
9,
873-879.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

