 |
PDBsum entry 2nwb

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
2nwb
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Crystal structure of a putative 2,3-dioxygenase (so4414) from shewanella oneidensis in complex with ferric heme. Northeast structural genomics target sor52.
|
|
Structure:
|
 |
Conserved domain protein. Chain: a, b. Synonym: putative 2,3-dioxygenase. Engineered: yes
|
|
Source:
|
 |
Shewanella oneidensis. Organism_taxid: 211586. Strain: mr-1. Gene: so_4414. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
2.40Å
|
R-factor:
|
0.217
|
R-free:
|
0.225
|
|
|
Authors:
|
 |
F.Forouhar,J.L.R.Anderson,C.G.Mowat,A.Hussain,J.Seetharaman, C.Bruckmann,S.J.Thackray,N.Khan,K.Cunningham,H.Janjua,L.Zhao,R.Xiao, L.C.Ma,J.Liu,M.C.Baran,T.B.Acton,B.Rost,G.T.Montelione,S.K.Champman, L.Tong,Northeast Structural Genomics Consortium (Nesg)
|
Key ref:

|
 |
F.Forouhar
et al.
(2007).
Molecular insights into substrate recognition and catalysis by tryptophan 2,3-dioxygenase.
Proc Natl Acad Sci U S A,
104,
473-478.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
14-Nov-06
|
Release date:
|
19-Dec-06
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q8E972
(Q8E972_SHEON) -
Tryptophan 2,3-dioxygenase KynA from Shewanella oneidensis (strain MR-1)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
392 a.a.
379 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.13.11.11
- tryptophan 2,3-dioxygenase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Tryptophan Catabolism
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-tryptophan + O2 = N-formyl-L-kynurenine
|
 |
 |
 |
 |
 |
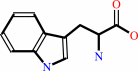 L-tryptophan
L-tryptophan
|
+
|
 O2
O2
|
=
|
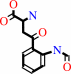 N-formyl-L-kynurenine
N-formyl-L-kynurenine
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Heme
|
 |
 |
 |
 |
 |
Heme
Bound ligand (Het Group name =
HEM)
matches with 95.45% similarity
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Proc Natl Acad Sci U S A
104:473-478
(2007)
|
|
PubMed id:
|
|
|
|

|
| |
|
Molecular insights into substrate recognition and catalysis by tryptophan 2,3-dioxygenase.
|
|
F.Forouhar,
J.L.Anderson,
C.G.Mowat,
S.M.Vorobiev,
A.Hussain,
M.Abashidze,
C.Bruckmann,
S.J.Thackray,
J.Seetharaman,
T.Tucker,
R.Xiao,
L.C.Ma,
L.Zhao,
T.B.Acton,
G.T.Montelione,
S.K.Chapman,
L.Tong.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Tryptophan 2,3-dioxygenase (TDO) and indoleamine 2,3-dioxygenase (IDO)
constitute an important, yet relatively poorly understood, family of
heme-containing enzymes. Here, we report extensive structural and biochemical
studies of the Xanthomonas campestris TDO and a related protein SO4414 from
Shewanella oneidensis, including the structure at 1.6-A resolution of the
catalytically active, ferrous form of TDO in a binary complex with the substrate
L-Trp. The carboxylate and ammonium moieties of tryptophan are recognized by
electrostatic and hydrogen-bonding interactions with the enzyme and a propionate
group of the heme, thus defining the L-stereospecificity. A second, possibly
allosteric, L-Trp-binding site is present at the tetramer interface. The sixth
coordination site of the heme-iron is vacant, providing a dioxygen-binding site
that would also involve interactions with the ammonium moiety of L-Trp and the
amide nitrogen of a glycine residue. The indole ring is positioned correctly for
oxygenation at the C2 and C3 atoms. The active site is fully formed only in the
binary complex, and biochemical experiments confirm this induced-fit behavior of
the enzyme. The active site is completely devoid of water during catalysis,
which is supported by our electrochemical studies showing significant
stabilization of the enzyme upon substrate binding.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 2.
Fig. 2. The structure of TDO. (a) Schematic representation
of the structure of the monomer of X. campestris TDO. The  -helices
are shown in yellow and labeled. Heme is shown in gray, and
L-Trp is shown in orange (labeled W). The water molecule is
shown as a red sphere (labeled wat). (b) Schematic
representation of the tetramer of X. campestris TDO. The four
monomers are colored in yellow, cyan, violet, and green. Helices
in the tetramer interface are labeled. The Trp molecules in the
tetramer interface are also shown. Produced with Molscript (35)
and rendered with Raster3D (36). -helices
are shown in yellow and labeled. Heme is shown in gray, and
L-Trp is shown in orange (labeled W). The water molecule is
shown as a red sphere (labeled wat). (b) Schematic
representation of the tetramer of X. campestris TDO. The four
monomers are colored in yellow, cyan, violet, and green. Helices
in the tetramer interface are labeled. The Trp molecules in the
tetramer interface are also shown. Produced with Molscript (35)
and rendered with Raster3D (36).
|
 |
Figure 3.
Fig. 3. Molecular basis for substrate recognition by TDO.
(a) Final 2F[o]–F[c] electron density at 1.6-Å
resolution for heme, L-Trp, and a water in the active site.
Contoured at 1  . (b) Stereo drawing
showing the active site of X. campestris TDO in the binary
complex with L-Trp. The segment in cyan is from another monomer
of the tetramer. Hydrogen-bonding interactions are indicated
with dashed lines in magenta. (c) Overlay of the structures of
the free enzyme (in orchid) and the binary complex (yellow and
cyan) in the active-site region. Regions of conformational
differences are indicated with the red arrows. (d) Overlay of
the active-site region of the second monomer (in green) and that
of the first monomer (in yellow). Only the side-chain atoms of
Trp are shown in the second monomer (in magenta). (e) Final
2F[o]–F[c] electron density at 1.6-Å resolution for
heme, L-Trp, and a water in the active site of the second TDO
molecule in the crystal. Contoured at 1 . (b) Stereo drawing
showing the active site of X. campestris TDO in the binary
complex with L-Trp. The segment in cyan is from another monomer
of the tetramer. Hydrogen-bonding interactions are indicated
with dashed lines in magenta. (c) Overlay of the structures of
the free enzyme (in orchid) and the binary complex (yellow and
cyan) in the active-site region. Regions of conformational
differences are indicated with the red arrows. (d) Overlay of
the active-site region of the second monomer (in green) and that
of the first monomer (in yellow). Only the side-chain atoms of
Trp are shown in the second monomer (in magenta). (e) Final
2F[o]–F[c] electron density at 1.6-Å resolution for
heme, L-Trp, and a water in the active site of the second TDO
molecule in the crystal. Contoured at 1  . Two conformations for
the main chain atoms are shown, but neither fit the density
well. For the stereo version of c and d, please see SI Fig. 7.
Produced with Molscript (35) and rendered with Raster3D (36). . Two conformations for
the main chain atoms are shown, but neither fit the density
well. For the stereo version of c and d, please see SI Fig. 7.
Produced with Molscript (35) and rendered with Raster3D (36).
|
 |
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
E.Olsson,
A.Martinez,
K.Teigen,
and
V.R.Jensen
(2011).
Formation of the iron-oxo hydroxylating species in the catalytic cycle of aromatic amino acid hydroxylases.
|
| |
Chemistry,
17,
3746-3758.
|
 |
|
|
|
|
 |
H.J.Yuasa,
and
H.J.Ball
(2011).
Molecular evolution and characterization of fungal indoleamine 2,3-dioxygenases.
|
| |
J Mol Evol,
72,
160-168.
|
 |
|
|
|
|
 |
J.E.Voss,
S.W.Scally,
N.L.Taylor,
S.C.Atkinson,
M.D.Griffin,
C.A.Hutton,
M.W.Parker,
M.R.Alderton,
J.A.Gerrard,
R.C.Dobson,
C.Dogovski,
and
M.A.Perugini
(2010).
Substrate-mediated stabilization of a tetrameric drug target reveals Achilles heel in anthrax.
|
| |
J Biol Chem,
285,
5188-5195.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
L.Capece,
A.Lewis-Ballester,
D.Batabyal,
N.Di Russo,
S.R.Yeh,
D.A.Estrin,
and
M.A.Marti
(2010).
The first step of the dioxygenation reaction carried out by tryptophan dioxygenase and indoleamine 2,3-dioxygenase as revealed by quantum mechanical/molecular mechanical studies.
|
| |
J Biol Inorg Chem,
15,
811-823.
|
 |
|
|
|
|
 |
L.Capece,
M.Arrar,
A.E.Roitberg,
S.R.Yeh,
M.A.Marti,
and
D.A.Estrin
(2010).
Substrate stereo-specificity in tryptophan dioxygenase and indoleamine 2,3-dioxygenase.
|
| |
Proteins,
78,
2961-2972.
|
 |
|
|
|
|
 |
R.M.Davydov,
N.Chauhan,
S.J.Thackray,
J.L.Anderson,
N.D.Papadopoulou,
C.G.Mowat,
S.K.Chapman,
E.L.Raven,
and
B.M.Hoffman
(2010).
Probing the ternary complexes of indoleamine and tryptophan 2,3-dioxygenases by cryoreduction EPR and ENDOR spectroscopy.
|
| |
J Am Chem Soc,
132,
5494-5500.
|
 |
|
|
|
|
 |
A.Lewis-Ballester,
D.Batabyal,
T.Egawa,
C.Lu,
Y.Lin,
M.A.Marti,
L.Capece,
D.A.Estrin,
and
S.R.Yeh
(2009).
Evidence for a ferryl intermediate in a heme-based dioxygenase.
|
| |
Proc Natl Acad Sci U S A,
106,
17371-17376.
|
 |
|
|
|
|
 |
E.Fukumura,
H.Sugimoto,
Y.Misumi,
T.Ogura,
and
Y.Shiro
(2009).
Cooperative binding of L-trp to human tryptophan 2,3-dioxygenase: resonance Raman spectroscopic analysis.
|
| |
J Biochem,
145,
505-515.
|
 |
|
|
|
|
 |
E.Nickel,
K.Nienhaus,
C.Lu,
S.R.Yeh,
and
G.U.Nienhaus
(2009).
Ligand and substrate migration in human indoleamine 2,3-dioxygenase.
|
| |
J Biol Chem,
284,
31548-31554.
|
 |
|
|
|
|
 |
A.Sheoran,
A.King,
A.Velasco,
J.M.Pero,
and
S.Garneau-Tsodikova
(2008).
Characterization of TioF, a tryptophan 2,3-dioxygenase involved in 3-hydroxyquinaldic acid formation during thiocoraline biosynthesis.
|
| |
Mol Biosyst,
4,
622-628.
|
 |
|
|
|
|
 |
S.J.Thackray,
C.G.Mowat,
and
S.K.Chapman
(2008).
Exploring the mechanism of tryptophan 2,3-dioxygenase.
|
| |
Biochem Soc Trans,
36,
1120-1123.
|
 |
|
|
|
|
 |
F.Delaspre,
C.G.Nieto Peñalver,
O.Saurel,
P.Kiefer,
E.Gras,
A.Milon,
C.Boucher,
S.Genin,
and
J.A.Vorholt
(2007).
The Ralstonia solanacearum pathogenicity regulator HrpB induces 3-hydroxy-oxindole synthesis.
|
| |
Proc Natl Acad Sci U S A,
104,
15870-15875.
|
 |
|
|
|
|
 |
F.Forouhar,
A.Kuzin,
J.Seetharaman,
I.Lee,
W.Zhou,
M.Abashidze,
Y.Chen,
W.Yong,
H.Janjua,
Y.Fang,
D.Wang,
K.Cunningham,
R.Xiao,
T.B.Acton,
E.Pichersky,
D.F.Klessig,
C.W.Porter,
G.T.Montelione,
and
L.Tong
(2007).
Functional insights from structural genomics.
|
| |
J Struct Funct Genomics,
8,
37-44.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
H.J.Yuasa,
M.Takubo,
A.Takahashi,
T.Hasegawa,
H.Noma,
and
T.Suzuki
(2007).
Evolution of vertebrate indoleamine 2,3-dioxygenases.
|
| |
J Mol Evol,
65,
705-714.
|
 |
|
|
|
|
 |
W.A.Hendrickson
(2007).
Impact of structures from the protein structure initiative.
|
| |
Structure,
15,
1528-1529.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

