 |
PDBsum entry 1tm0

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Isomerase
|
 |
|
Title:
|
 |
Crystal structure of the putative proline racemase from brucella melitensis, northeast structural genomics target lr31
|
|
Structure:
|
 |
Proline racemase. Chain: a, b. Engineered: yes
|
|
Source:
|
 |
Brucella melitensis. Organism_taxid: 224914. Strain: 16m. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
2.80Å
|
R-factor:
|
0.236
|
R-free:
|
0.312
|
|
|
Authors:
|
 |
F.Forouhar,Y.Chen,R.Xiao,C.K.Ho,L.-C.Ma,B.Cooper,T.B.Acton, G.T.Montelione,J.F.Hunt,L.Tong,Northeast Structural Genomics Consortium (Nesg)
|
|
Key ref:
|
 |
F.Forouhar
et al.
(2007).
Functional insights from structural genomics.
J Struct Funct Genomics,
8,
37-44.
PubMed id:
|
 |
|
Date:
|
 |
|
10-Jun-04
|
Release date:
|
29-Jun-04
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q8YFD6
(Y1586_BRUME) -
Protein BMEI1586 from Brucella melitensis biotype 1 (strain ATCC 23456 / CCUG 17765 / NCTC 10094 / 16M)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
342 a.a.
311 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.5.1.1.8
- 4-hydroxyproline epimerase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
4-Hydroxyproline Epimerase
|
 |
 |
 |
 |
 |
Reaction:
|
 |
trans-4-hydroxy-L-proline = cis-4-hydroxy-D-proline
|
 |
 |
 |
 |
 |
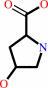 trans-4-hydroxy-L-proline
trans-4-hydroxy-L-proline
|
=
|
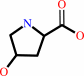 cis-4-hydroxy-D-proline
cis-4-hydroxy-D-proline
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
J Struct Funct Genomics
8:37-44
(2007)
|
|
PubMed id:
|
|
|
|

|
| |
|
Functional insights from structural genomics.
|
|
F.Forouhar,
A.Kuzin,
J.Seetharaman,
I.Lee,
W.Zhou,
M.Abashidze,
Y.Chen,
W.Yong,
H.Janjua,
Y.Fang,
D.Wang,
K.Cunningham,
R.Xiao,
T.B.Acton,
E.Pichersky,
D.F.Klessig,
C.W.Porter,
G.T.Montelione,
L.Tong.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Structural genomics efforts have produced structural information, either
directly or by modeling, for thousands of proteins over the past few years.
While many of these proteins have known functions, a large percentage of them
have not been characterized at the functional level. The structural information
has provided valuable functional insights on some of these proteins, through
careful structural analyses, serendipity, and structure-guided functional
screening. Some of the success stories based on structures solved at the
Northeast Structural Genomics Consortium (NESG) are reported here. These include
a novel methyl salicylate esterase with important role in plant innate immunity,
a novel RNA methyltransferase (H. influenzae yggJ (HI0303)), a novel
spermidine/spermine N-acetyltransferase (B. subtilis PaiA), a novel
methyltransferase or AdoMet binding protein (A. fulgidus AF_0241), an
ATP:cob(I)alamin adenosyltransferase (B. subtilis YvqK), a novel carboxysome
pore (E. coli EutN), a proline racemase homolog with a disrupted active site (B.
melitensis BME11586), an FMN-dependent enzyme (S. pneumoniae SP_1951), and a
12-stranded beta-barrel with a novel fold (V. parahaemolyticus VPA1032).

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
C.A.Kerfeld,
S.Heinhorst,
and
G.C.Cannon
(2010).
Bacterial microcompartments.
|
| |
Annu Rev Microbiol,
64,
391-408.
|
 |
|
|
|
|
 |
D.A.Garsin
(2010).
Ethanolamine utilization in bacterial pathogens: roles and regulation.
|
| |
Nat Rev Microbiol,
8,
290-295.
|
 |
|
|
|
|
 |
T.O.Yeates,
C.S.Crowley,
and
S.Tanaka
(2010).
Bacterial microcompartment organelles: protein shell structure and evolution.
|
| |
Annu Rev Biophys,
39,
185-205.
|
 |
|
|
|
|
 |
T.Triplet,
M.D.Shortridge,
M.A.Griep,
J.L.Stark,
R.Powers,
and
P.Revesz
(2010).
PROFESS: a PROtein function, evolution, structure and sequence database.
|
| |
Database (Oxford),
2010,
baq011.
|
 |
|
|
|
|
 |
M.Beeby,
T.A.Bobik,
and
T.O.Yeates
(2009).
Exploiting genomic patterns to discover new supramolecular protein assemblies.
|
| |
Protein Sci,
18,
69-79.
|
 |
|
|
|
|
 |
M.Sagermann,
A.Ohtaki,
and
K.Nikolakakis
(2009).
Crystal structure of the EutL shell protein of the ethanolamine ammonia lyase microcompartment.
|
| |
Proc Natl Acad Sci U S A,
106,
8883-8887.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
E.M.Sampson,
and
T.A.Bobik
(2008).
Microcompartments for B12-dependent 1,2-propanediol degradation provide protection from DNA and cellular damage by a reactive metabolic intermediate.
|
| |
J Bacteriol,
190,
2966-2971.
|
 |
|
|
|
|
 |
J.B.Parsons,
S.D.Dinesh,
E.Deery,
H.K.Leech,
A.A.Brindley,
D.Heldt,
S.Frank,
C.M.Smales,
H.Lünsdorf,
A.Rambach,
M.H.Gass,
A.Bleloch,
K.J.McClean,
A.W.Munro,
S.E.Rigby,
M.J.Warren,
and
M.B.Prentice
(2008).
Biochemical and structural insights into bacterial organelle form and biogenesis.
|
| |
J Biol Chem,
283,
14366-14375.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

