 |
PDBsum entry 2nox

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
2nox
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Crystal structure of tryptophan 2,3-dioxygenase from ralstonia metallidurans
|
|
Structure:
|
 |
Tryptophan 2,3-dioxygenase. Chain: a, b, c, d, e, f, g, h, i, j, k, l, m, n, o, p. Ec: 1.13.11.11
|
|
Source:
|
 |
Cupriavidus metallidurans. Organism_taxid: 119219
|
|
Resolution:
|
 |
|
2.40Å
|
R-factor:
|
0.210
|
R-free:
|
0.270
|
|
|
Authors:
|
 |
Y.Zhang,S.A.Kang,T.Mukherjee,S.Bale,B.R.Crane,T.P.Begley,S.E.Ealick
|
|
Key ref:
|
 |
Y.Zhang
et al.
(2007).
Crystal structure and mechanism of tryptophan 2,3-dioxygenase, a heme enzyme involved in tryptophan catabolism and in quinolinate biosynthesis.
Biochemistry,
46,
145-155.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
26-Oct-06
|
Release date:
|
19-Dec-06
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q1LK00
(T23O_CUPMC) -
Tryptophan 2,3-dioxygenase from Cupriavidus metallidurans (strain ATCC 43123 / DSM 2839 / NBRC 102507 / CH34)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
299 a.a.
261 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.13.11.11
- tryptophan 2,3-dioxygenase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Tryptophan Catabolism
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-tryptophan + O2 = N-formyl-L-kynurenine
|
 |
 |
 |
 |
 |
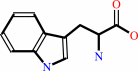 L-tryptophan
L-tryptophan
|
+
|
 O2
O2
|
=
|
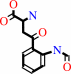 N-formyl-L-kynurenine
N-formyl-L-kynurenine
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Heme
|
 |
 |
 |
 |
 |
Heme
Bound ligand (Het Group name =
HEM)
matches with 95.45% similarity
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
46:145-155
(2007)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure and mechanism of tryptophan 2,3-dioxygenase, a heme enzyme involved in tryptophan catabolism and in quinolinate biosynthesis.
|
|
Y.Zhang,
S.A.Kang,
T.Mukherjee,
S.Bale,
B.R.Crane,
T.P.Begley,
S.E.Ealick.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The structure of tryptophan 2,3-dioxygenase (TDO) from Ralstonia metallidurans
was determined at 2.4 A. TDO catalyzes the irreversible oxidation of
l-tryptophan to N-formyl kynurenine, which is the initial step in tryptophan
catabolism. TDO is a heme-containing enzyme and is highly specific for its
substrate l-tryptophan. The structure is a tetramer with a heme cofactor bound
at each active site. The monomeric fold, as well as the heme binding site, is
similar to that of the large domain of indoleamine 2,3-dioxygenase, an enzyme
that catalyzes the same reaction except with a broader substrate tolerance.
Modeling of the putative (S)-tryptophan hydroperoxide intermediate into the
active site, as well as substrate analogue and mutagenesis studies, are
consistent with a Criegee mechanism for the reaction.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
L.Capece,
A.Lewis-Ballester,
D.Batabyal,
N.Di Russo,
S.R.Yeh,
D.A.Estrin,
and
M.A.Marti
(2010).
The first step of the dioxygenation reaction carried out by tryptophan dioxygenase and indoleamine 2,3-dioxygenase as revealed by quantum mechanical/molecular mechanical studies.
|
| |
J Biol Inorg Chem,
15,
811-823.
|
 |
|
|
|
|
 |
L.Capece,
M.Arrar,
A.E.Roitberg,
S.R.Yeh,
M.A.Marti,
and
D.A.Estrin
(2010).
Substrate stereo-specificity in tryptophan dioxygenase and indoleamine 2,3-dioxygenase.
|
| |
Proteins,
78,
2961-2972.
|
 |
|
|
|
|
 |
R.M.Davydov,
N.Chauhan,
S.J.Thackray,
J.L.Anderson,
N.D.Papadopoulou,
C.G.Mowat,
S.K.Chapman,
E.L.Raven,
and
B.M.Hoffman
(2010).
Probing the ternary complexes of indoleamine and tryptophan 2,3-dioxygenases by cryoreduction EPR and ENDOR spectroscopy.
|
| |
J Am Chem Soc,
132,
5494-5500.
|
 |
|
|
|
|
 |
U.Grohmann,
and
V.Bronte
(2010).
Control of immune response by amino acid metabolism.
|
| |
Immunol Rev,
236,
243-264.
|
 |
|
|
|
|
 |
A.Lewis-Ballester,
D.Batabyal,
T.Egawa,
C.Lu,
Y.Lin,
M.A.Marti,
L.Capece,
D.A.Estrin,
and
S.R.Yeh
(2009).
Evidence for a ferryl intermediate in a heme-based dioxygenase.
|
| |
Proc Natl Acad Sci U S A,
106,
17371-17376.
|
 |
|
|
|
|
 |
A.Macchiarulo,
E.Camaioni,
R.Nuti,
and
R.Pellicciari
(2009).
Highlights at the gate of tryptophan catabolism: a review on the mechanisms of activation and regulation of indoleamine 2,3-dioxygenase (IDO), a novel target in cancer disease.
|
| |
Amino Acids,
37,
219-229.
|
 |
|
|
|
|
 |
S.Löb,
A.Königsrainer,
H.G.Rammensee,
G.Opelz,
and
P.Terness
(2009).
Inhibitors of indoleamine-2,3-dioxygenase for cancer therapy: can we see the wood for the trees?
|
| |
Nat Rev Cancer,
9,
445-452.
|
 |
|
|
|
|
 |
A.Sheoran,
A.King,
A.Velasco,
J.M.Pero,
and
S.Garneau-Tsodikova
(2008).
Characterization of TioF, a tryptophan 2,3-dioxygenase involved in 3-hydroxyquinaldic acid formation during thiocoraline biosynthesis.
|
| |
Mol Biosyst,
4,
622-628.
|
 |
|
|
|
|
 |
M.von Grotthuss,
D.Plewczynski,
G.Vriend,
and
L.Rychlewski
(2008).
3D-Fun: predicting enzyme function from structure.
|
| |
Nucleic Acids Res,
36,
W303-W307.
|
 |
|
|
|
|
 |
S.J.Thackray,
C.G.Mowat,
and
S.K.Chapman
(2008).
Exploring the mechanism of tryptophan 2,3-dioxygenase.
|
| |
Biochem Soc Trans,
36,
1120-1123.
|
 |
|
|
|
|
 |
H.J.Yuasa,
M.Takubo,
A.Takahashi,
T.Hasegawa,
H.Noma,
and
T.Suzuki
(2007).
Evolution of vertebrate indoleamine 2,3-dioxygenases.
|
| |
J Mol Evol,
65,
705-714.
|
 |
|
|
|
|
 |
S.R.Thomas,
A.C.Terentis,
H.Cai,
O.Takikawa,
A.Levina,
P.A.Lay,
M.Freewan,
and
R.Stocker
(2007).
Post-translational regulation of human indoleamine 2,3-dioxygenase activity by nitric oxide.
|
| |
J Biol Chem,
282,
23778-23787.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

