 |
PDBsum entry 2hgs

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Amine/carboxylate ligase
|
PDB id
|
|

|

|
2hgs
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.6.3.2.3
- glutathione synthase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
gamma-L-glutamyl-L-cysteine + glycine + ATP = glutathione + ADP + phosphate + H+
|
 |
 |
 |
 |
 |
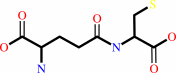 gamma-L-glutamyl-L-cysteine
gamma-L-glutamyl-L-cysteine
|
+
|
 glycine
glycine
|
+
|
 ATP
ATP
|
=
|
glutathione
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
ADP
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
 phosphate
phosphate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
EMBO J
18:3204-3213
(1999)
|
|
PubMed id:
|
|
|
|

|
| |
|
Molecular basis of glutathione synthetase deficiency and a rare gene permutation event.
|
|
G.Polekhina,
P.G.Board,
R.R.Gali,
J.Rossjohn,
M.W.Parker.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Glutathione synthetase (GS) catalyses the production of glutathione from
gamma-glutamylcysteine and glycine in an ATP-dependent manner. Malfunctioning of
GS results in disorders including metabolic acidosis, 5-oxoprolinuria,
neurological dysfunction, haemolytic anaemia and in some cases is probably
lethal. Here we report the crystal structure of human GS (hGS) at 2.1 A
resolution in complex with ADP, two magnesium ions, a sulfate ion and
glutathione. The structure indicates that hGS belongs to the recently identified
ATP-grasp superfamily, although it displays no detectable sequence identity with
other family members including its bacterial counterpart, Escherichia coli GS.
The difficulty in identifying hGS as a member of the family is due in part to a
rare gene permutation which has resulted in a circular shift of the conserved
secondary structure elements in hGS with respect to the other known ATP-grasp
proteins. Nevertheless, it appears likely that the enzyme shares the same
general catalytic mechanism as other ligases. The possibility of cyclic
permutations provides an insight into the evolution of this family and will
probably lead to the identification of new members. Mutations that lead to GS
deficiency have been mapped onto the structure, providing a molecular basis for
understanding their effects.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 3.
Figure 3 A ribbon representation of the dimer. The dimerization
unit of each monomer is shown in different colour and the lid
domains shown in mauve. The GS deficiency mutations are shown in
ball-and-stick. This figure was drawn with MOLSCRIPT (Kraulis,
1991).
|
 |
Figure 4.
Figure 4 Comparison of human and bacterial GS. (A)
Structure-based sequence alignment of hGS and ecGS. The
secondary structure and residue numbering of hGS are shown above
the alignment and the ecGS numbering below it. The secondary
structure of the conserved structural core elements of the
ATP-grasp superfamily are coloured as follows: the N-terminal
domain is red, the middle, lid domain is green, the C-terminal
domain is blue and the linker region (to the lid domain) in
orange. Additional secondary structures found in hGS are shown
in gray. Strictly conserved residues are highlighted in darkened
boxes and invariant residues of GS enzymes from eukaryotic
organisms are designated by asterisks. The complimentary
mutations and cis-peptide regions discussed in the text are
highlighted in the open boxes. The figure was produced using
ALSCRIPT (Barton, 1993). (B and C) Topological diagrams
highlighting the major structural elements of hGS and ecGS. The
colours refer to the structural cores conserved throughout the
ATP-grasp superfamily. (D and E) Ribbon diagrams of hGS and ecGS
using the same colour coding as in (A). The ribbon figures were
drawn with MOLSCRIPT (Kraulis, 1991).
|
 |
|
|

|
| |
The above figures are
reprinted
from an Open Access publication published by Macmillan Publishers Ltd:
EMBO J
(1999,
18,
3204-3213)
copyright 1999.
|
|
| |
Figures were
selected
by the author.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
I.Wohlers,
F.S.Domingues,
and
G.W.Klau
(2010).
Towards optimal alignment of protein structure distance matrices.
|
| |
Bioinformatics,
26,
2273-2280.
|
 |
|
|
|
|
 |
M.V.Omelchenko,
M.Y.Galperin,
Y.I.Wolf,
and
E.V.Koonin
(2010).
Non-homologous isofunctional enzymes: a systematic analysis of alternative solutions in enzyme evolution.
|
| |
Biol Direct,
5,
31.
|
 |
|
|
|
|
 |
P.K.Fyfe,
M.S.Alphey,
and
W.N.Hunter
(2010).
Structure of Trypanosoma brucei glutathione synthetase: domain and loop alterations in the catalytic cycle of a highly conserved enzyme.
|
| |
Mol Biochem Parasitol,
170,
93-99.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.Deville,
J.Rey,
and
M.Chabbert
(2008).
Comprehensive analysis of the helix-X-helix motif in soluble proteins.
|
| |
Proteins,
72,
115-135.
|
 |
|
|
|
|
 |
W.C.Lo,
and
P.C.Lyu
(2008).
CPSARST: an efficient circular permutation search tool applied to the detection of novel protein structural relationships.
|
| |
Genome Biol,
9,
R11.
|
 |
|
|
|
|
 |
A.Abyzov,
and
V.A.Ilyin
(2007).
A comprehensive analysis of non-sequential alignments between all protein structures.
|
| |
BMC Struct Biol,
7,
78.
|
 |
|
|
|
|
 |
E.Ristoff,
and
A.Larsson
(2007).
Inborn errors in the metabolism of glutathione.
|
| |
Orphanet J Rare Dis,
2,
16.
|
 |
|
|
|
|
 |
G.Ausiello,
D.Peluso,
A.Via,
and
M.Helmer-Citterich
(2007).
Local comparison of protein structures highlights cases of convergent evolution in analogous functional sites.
|
| |
BMC Bioinformatics,
8,
S24.
|
 |
|
|
|
|
 |
K.Herrera,
R.E.Cahoon,
S.Kumaran,
and
J.Jez
(2007).
Reaction mechanism of glutathione synthetase from Arabidopsis thaliana: site-directed mutagenesis of active site residues.
|
| |
J Biol Chem,
282,
17157-17165.
|
 |
|
|
|
|
 |
N.Nagano,
T.Noguchi,
and
Y.Akiyama
(2007).
Systematic comparison of catalytic mechanisms of hydrolysis and transfer reactions classified in the EzCatDB database.
|
| |
Proteins,
66,
147-159.
|
 |
|
|
|
|
 |
B.Vergauwen,
D.De Vos,
and
J.J.Van Beeumen
(2006).
Characterization of the bifunctional gamma-glutamate-cysteine ligase/glutathione synthetase (GshF) of Pasteurella multocida.
|
| |
J Biol Chem,
281,
4380-4394.
|
 |
|
|
|
|
 |
C.H.Pai,
B.Y.Chiang,
T.P.Ko,
C.C.Chou,
C.M.Chong,
F.J.Yen,
S.Chen,
J.K.Coward,
A.H.Wang,
and
C.H.Lin
(2006).
Dual binding sites for translocation catalysis by Escherichia coli glutathionylspermidine synthetase.
|
| |
EMBO J,
25,
5970-5982.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.E.Fraser,
K.Hayakawa,
M.S.Hume,
D.G.Ryan,
and
E.R.Brownie
(2006).
Interactions of GTP with the ATP-grasp domain of GTP-specific succinyl-CoA synthetase.
|
| |
J Biol Chem,
281,
11058-11065.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Comini,
U.Menge,
J.Wissing,
and
L.Flohé
(2005).
Trypanothione synthesis in crithidia revisited.
|
| |
J Biol Chem,
280,
6850-6860.
|
 |
|
|
|
|
 |
R.Njålsson,
and
S.Norgren
(2005).
Physiological and pathological aspects of GSH metabolism.
|
| |
Acta Paediatr,
94,
132-137.
|
 |
|
|
|
|
 |
J.M.Jez,
and
R.E.Cahoon
(2004).
Kinetic mechanism of glutathione synthetase from Arabidopsis thaliana.
|
| |
J Biol Chem,
279,
42726-42731.
|
 |
|
|
|
|
 |
H.Li,
H.Xu,
D.E.Graham,
and
R.H.White
(2003).
Glutathione synthetase homologs encode alpha-L-glutamate ligases for methanogenic coenzyme F420 and tetrahydrosarcinapterin biosyntheses.
|
| |
Proc Natl Acad Sci U S A,
100,
9785-9790.
|
 |
|
|
|
|
 |
N.Phlippen,
K.Hoffmann,
R.Fischer,
K.Wolf,
and
M.Zimmermann
(2003).
The glutathione synthetase of Schizosaccharomyces pombe is synthesized as a homodimer but retains full activity when present as a heterotetramer.
|
| |
J Biol Chem,
278,
40152-40161.
|
 |
|
|
|
|
 |
C.H.Williams,
T.J.Stillman,
V.V.Barynin,
S.E.Sedelnikova,
Y.Tang,
J.Green,
J.R.Guest,
and
P.J.Artymiuk
(2002).
E. coli aconitase B structure reveals a HEAT-like domain with implications for protein-protein recognition.
|
| |
Nat Struct Biol,
9,
447-452.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.D.Copley,
and
J.K.Dhillon
(2002).
Lateral gene transfer and parallel evolution in the history of glutathione biosynthesis genes.
|
| |
Genome Biol,
3,
research0025.
|
 |
|
|
|
|
 |
J.Jung,
and
B.Lee
(2001).
Circularly permuted proteins in the protein structure database.
|
| |
Protein Sci,
10,
1881-1886.
|
 |
|
|
|
|
 |
K.D.Yang,
and
H.R.Hill
(2001).
Granulocyte function disorders: aspects of development, genetics and management.
|
| |
Pediatr Infect Dis J,
20,
889-900.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

