 |
PDBsum entry 2fpx

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
Chains A, B:
E.C.3.1.3.15
- histidinol-phosphatase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Histidine Biosynthesis (late stages)
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-histidinol phosphate + H2O = L-histidinol + phosphate
|
 |
 |
 |
 |
 |
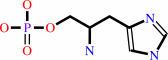 L-histidinol phosphate
L-histidinol phosphate
|
+
|
H2O
|
=
|
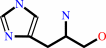 L-histidinol
L-histidinol
|
+
|
 phosphate
phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
Chains A, B:
E.C.4.2.1.19
- imidazoleglycerol-phosphate dehydratase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
|
 |
 |
 |
 |
 |
Reaction:
|
 |
D-erythro-1-(imidazol-4-yl)glycerol 3-phosphate = 3-(imidazol-4-yl)-2- oxopropyl phosphate + H2O
|
 |
 |
 |
 |
 |
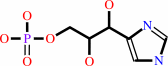 D-erythro-1-(imidazol-4-yl)glycerol 3-phosphate
D-erythro-1-(imidazol-4-yl)glycerol 3-phosphate
|
=
|
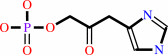 3-(imidazol-4-yl)-2- oxopropyl phosphate
3-(imidazol-4-yl)-2- oxopropyl phosphate
|
+
|
H2O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
281:37930-37941
(2006)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural snapshots of Escherichia coli histidinol phosphate phosphatase along the reaction pathway.
|
|
E.S.Rangarajan,
A.Proteau,
J.Wagner,
M.N.Hung,
A.Matte,
M.Cygler.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
HisB from Escherichia coli is a bifunctional enzyme catalyzing the sixth and
eighth steps of l-histidine biosynthesis. The N-terminal domain (HisB-N)
possesses histidinol phosphate phosphatase activity, and its crystal structure
shows a single domain with fold similarity to the haloacid dehalogenase (HAD)
enzyme family. HisB-N forms dimers in the crystal and in solution. The structure
shows the presence of a structural Zn(2+) ion stabilizing the conformation of an
extended loop. Two metal binding sites were also identified in the active site.
Their presence was further confirmed by isothermal titration calorimetry. HisB-N
is active in the presence of Mg(2+), Mn(2+), Co(2+), or Zn(2+), but Ca(2+) has
an inhibitory effect. We have determined structures of several intermediate
states corresponding to snapshots along the reaction pathway, including that of
the phosphoaspartate intermediate. A catalytic mechanism, different from that
described for other HAD enzymes, is proposed requiring the presence of the
second metal ion not found in the active sites of previously characterized HAD
enzymes, to complete the second half-reaction. The proposed mechanism is
reminiscent of two-Mg(2+) ion catalysis utilized by DNA and RNA polymerases and
many nucleases. The structure also provides an explanation for the inhibitory
effect of Ca(2+).

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 5.
FIGURE 5. a, states along the reaction pathway based on
determined structures. I, the initial state with site 1 occupied
by Mg^2+; II, HisB-N with histidinol phosphate substrate modeled
on the structure of histidinol complex; III, phosphoaspartate
intermediate with Mg^2+ occupying sites 1 and 2; IV, release of
phosphate and Mg^2+ from site 2; b, superposition of the active
site residues of HisB-N (gray) with E. coli AphA (orange). Red
spheres represent water molecules; violet spheres show metal
binding sites. The orientations of Asp^12 and its equivalent in
AphA are different. Asp^12 is stabilized by hydrogen bonds
(blue) to Arg^11, site 2, and bridging water W4. Asp^46 of AphA
is hydrogen-bonded (orange) to Arg^114, approaching from the
opposite direction to Arg^11 of HisB-N, and to the of phosphate
oxygen in the position of water W1. Only HisB residues are
labeled.
|
 |
Figure 6.
FIGURE 6. Changes in the HisB-N substrate binding region
along the reaction pathway shown in a surface representation.
The residues that undergo conformational changes due to binding
of the substrate and/or metal ions, namely Glu^18, Phe^23, and
Arg^132 are shown as sticks under semitransparent surface. All
structures are shown in the same orientation. a,
HisB-N·Mg; b, HisB-N·Mg/histidinol; c,
HisB-N·Ca/pAsp; and d, HisB-N·Mg/sulfate.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2006,
281,
37930-37941)
copyright 2006.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
H.H.Nguyen,
L.Wang,
H.Huang,
E.Peisach,
D.Dunaway-Mariano,
and
K.N.Allen
(2010).
Structural determinants of substrate recognition in the HAD superfamily member D-glycero-D-manno-heptose-1,7-bisphosphate phosphatase (GmhB) .
|
| |
Biochemistry,
49,
1082-1092.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
L.Wang,
H.Huang,
H.H.Nguyen,
K.N.Allen,
P.S.Mariano,
and
D.Dunaway-Mariano
(2010).
Divergence of biochemical function in the HAD superfamily: D-glycero-D-manno-heptose-1,7-bisphosphate phosphatase (GmhB).
|
| |
Biochemistry,
49,
1072-1081.
|
 |
|
|
|
|
 |
K.N.Allen,
and
D.Dunaway-Mariano
(2009).
Markers of fitness in a successful enzyme superfamily.
|
| |
Curr Opin Struct Biol,
19,
658-665.
|
 |
|
|
|
|
 |
H.S.Lee,
Y.Cho,
J.H.Lee,
and
S.G.Kang
(2008).
Novel monofunctional histidinol-phosphate phosphatase of the DDDD superfamily of phosphohydrolases.
|
| |
J Bacteriol,
190,
2629-2632.
|
 |
|
|
|
|
 |
O.Okhrimenko,
and
I.Jelesarov
(2008).
A survey of the year 2006 literature on applications of isothermal titration calorimetry.
|
| |
J Mol Recognit,
21,
1.
|
 |
|
|
|
|
 |
R.Schnell,
D.Agren,
and
G.Schneider
(2008).
1.9 A structure of the signal receiver domain of the putative response regulator NarL from Mycobacterium tuberculosis.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
64,
1096-1100.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.Matte,
Z.Jia,
S.Sunita,
J.Sivaraman,
and
M.Cygler
(2007).
Insights into the biology of Escherichia coli through structural proteomics.
|
| |
J Struct Funct Genomics,
8,
45-55.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |
|

