 |
PDBsum entry 2fc2

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
2fc2
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.14.14.47
- nitric-oxide synthase (flavodoxin).
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
3 reduced [flavodoxin] + 2 L-arginine + 4 O2 = 3 oxidized [flavodoxin] + 2 L-citrulline + 2 nitric oxide + 4 H2O + 5 H+
|
 |
 |
 |
 |
 |
3
×
reduced [flavodoxin]
Bound ligand (Het Group name = )
matches with 92.31% similarity
|
+
|
 2
×
L-arginine
2
×
L-arginine
|
+
|
 4
×
O2
4
×
O2
|
=
|
3
×
oxidized [flavodoxin]
|
+
|
 2
×
L-citrulline
2
×
L-citrulline
|
+
|
 2
×
nitric oxide
2
×
nitric oxide
|
+
|
4
×
H2O
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
5
×
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
5,6,7,8-tetrahydrobiopterin; Ferriheme b
|
 |
 |
 |
 |
 |
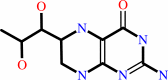 5,6,7,8-tetrahydrobiopterin
5,6,7,8-tetrahydrobiopterin
|
Ferriheme b
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
45:2537-2544
(2006)
|
|
PubMed id:
|
|
|
|

|
| |
|
Nitrosyl-heme structures of Bacillus subtilis nitric oxide synthase have implications for understanding substrate oxidation.
|
|
K.Pant,
B.R.Crane.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The crystal structures of nitrosyl-heme complexes of a prokaryotic nitric oxide
synthase (NOS) from Bacillus subtilis (bsNOS) reveal changes in active-site
hydrogen bonding in the presence of the intermediate N(omega)-hydroxy-l-arginine
(NOHA) compared to the substrate l-arginine (l-Arg). Correlating with a
Val-to-Ile residue substitution in the bsNOS heme pocket, the Fe(II)-NO complex
with both l-Arg and NOHA is more bent than the Fe(II)-NO, l-Arg complex of
mammalian eNOS [Li, H., Raman, C. S., Martasek, P., Masters, B. S. S., and
Poulos, T. L. (2001) Biochemistry 40, 5399-5406]. Structures of the Fe(III)-NO
complex with NOHA show a nearly linear nitrosyl group, and in one subunit,
partial nitrosation of bound NOHA. In the Fe(II)-NO complexes, the protonated
NOHA N(omega) atom forms a short hydrogen bond with the heme-coordinated NO
nitrogen, but active-site water molecules are out of hydrogen bonding range with
the distal NO oxygen. In contrast, the l-Arg guanidinium interacts more weakly
and equally with both NO atoms, and an active-site water molecule hydrogen bonds
to the distal NO oxygen. This difference in hydrogen bonding to the nitrosyl
group by the two substrates indicates that interactions provided by NOHA may
preferentially stabilize an electrophilic peroxo-heme intermediate in the second
step of NOS catalysis.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.V.Soldatova,
M.Ibrahim,
J.S.Olson,
R.S.Czernuszewicz,
and
T.G.Spiro
(2010).
New light on NO bonding in Fe(III) heme proteins from resonance raman spectroscopy and DFT modeling.
|
| |
J Am Chem Soc,
132,
4614-4625.
|
 |
|
|
|
|
 |
B.R.Crane,
J.Sudhamsu,
and
B.A.Patel
(2010).
Bacterial nitric oxide synthases.
|
| |
Annu Rev Biochem,
79,
445-470.
|
 |
|
|
|
|
 |
C.Giroud,
M.Moreau,
T.A.Mattioli,
V.Balland,
J.L.Boucher,
Y.Xu-Li,
D.J.Stuehr,
and
J.Santolini
(2010).
Role of arginine guanidinium moiety in nitric-oxide synthase mechanism of oxygen activation.
|
| |
J Biol Chem,
285,
7233-7245.
|
 |
|
|
|
|
 |
J.Sudhamsu,
and
B.R.Crane
(2009).
Bacterial nitric oxide synthases: what are they good for?
|
| |
Trends Microbiol,
17,
212-218.
|
 |
|
|
|
|
 |
R.Wang,
M.A.Camacho-Fernandez,
W.Xu,
J.Zhang,
and
L.Li
(2009).
Neutral and reduced Roussin's red salt ester [Fe(2)(mu-RS)(2)(NO)(4)] (R = n-Pr, t-Bu, 6-methyl-2-pyridyl and 4,6-dimethyl-2-pyrimidyl): synthesis, X-ray crystal structures, spectroscopic, electrochemical and density functional theoretical investigations.
|
| |
Dalton Trans,
(),
777-786.
|
 |
|
|
|
|
 |
I.Gusarov,
M.Starodubtseva,
Z.Q.Wang,
L.McQuade,
S.J.Lippard,
D.J.Stuehr,
and
E.Nudler
(2008).
Bacterial nitric-oxide synthases operate without a dedicated redox partner.
|
| |
J Biol Chem,
283,
13140-13147.
|
 |
|
|
|
|
 |
D.P.Linder,
and
K.R.Rodgers
(2007).
Computational modeling of factors that modulate the unique FeNO bonding in {FeNO}(6) heme-thiolate model complexes.
|
| |
J Biol Inorg Chem,
12,
721-731.
|
 |
|
|
|
|
 |
F.J.Chartier,
and
M.Couture
(2007).
Substrate-specific interactions with the heme-bound oxygen molecule of nitric-oxide synthase.
|
| |
J Biol Chem,
282,
20877-20886.
|
 |
|
|
|
|
 |
H.Li,
J.Igarashi,
J.Jamal,
W.Yang,
and
T.L.Poulos
(2006).
Structural studies of constitutive nitric oxide synthases with diatomic ligands bound.
|
| |
J Biol Inorg Chem,
11,
753-768.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
N.Xu,
D.R.Powell,
L.Cheng,
and
G.B.Richter-Addo
(2006).
The first structurally characterized nitrosyl heme thiolate model complex.
|
| |
Chem Commun (Camb),
(),
2030-2032.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

