 |
PDBsum entry 2dig

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
DNA binding protein
|
PDB id
|
|

|

|
2dig
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.3.1.70
- Delta(14)-sterol reductase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Sterol ring B, C, D modification
|
 |
 |
 |
 |
 |
Reaction:
|
 |
4,4-dimethyl-5alpha-cholesta-8,24-dien-3beta-ol + NADP+ = 4,4-dimethyl- 5alpha-cholesta-8,14,24-trien-3beta-ol + NADPH + H+
|
 |
 |
 |
 |
 |
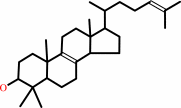 4,4-dimethyl-5alpha-cholesta-8,24-dien-3beta-ol
4,4-dimethyl-5alpha-cholesta-8,24-dien-3beta-ol
|
+
|
 NADP(+)
NADP(+)
|
=
|
4,4-dimethyl- 5alpha-cholesta-8,14,24-trien-3beta-ol
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

