
|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Crystal structure of the carbinolamine intermediate in the reductive half-reaction of aromatic amine dehydrogenase (aadh) with tryptamine. Monoclinic form
|
|
Structure:
|
 |
Aromatic amine dehydrogenase. Chain: d, h. Fragment: residues 48-182. Aromatic amine dehydrogenase. Chain: a, b. Fragment: residues 73-433. Ec: 1.4.99.4
|
|
Source:
|
 |
Alcaligenes faecalis. Organism_taxid: 511. Strain: ifo 14479. Strain: ifo 14479
|
|
Biol. unit:
|
 |
Tetramer (from
)
|
|
Resolution:
|
 |
|
1.45Å
|
R-factor:
|
0.169
|
R-free:
|
0.200
|
|
|
Authors:
|
 |
L.Masgrau,A.Roujeinikova,L.O.Johannissen,P.Hothi,J.Basran, K.E.Ranaghan,A.J.Mulholland,M.J.Sutcliffe,N.S.Scrutton,D.Leys
|
Key ref:

|
 |
L.Masgrau
et al.
(2006).
Atomic description of an enzyme reaction dominated by proton tunneling.
Science,
312,
237-241.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
27-Jul-05
|
Release date:
|
25-Apr-06
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chains D, H, A, B:
E.C.1.4.9.2
- aralkylamine dehydrogenase (azurin).
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
an aralkylamine + 2 oxidized [azurin] + H2O = an aromatic aldehyde + 2 reduced [azurin] + NH4+ + 2 H+
|
 |
 |
 |
 |
 |
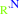 aralkylamine
aralkylamine
|
+
|
2
×
oxidized [azurin]
|
+
|
H2O
|
=
|
aromatic aldehyde
Bound ligand (Het Group name = )
matches with 61.54% similarity
|
+
|
2
×
reduced [azurin]
|
+
|
NH4(+)
|
+
|
2
×
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Science
312:237-241
(2006)
|
|
PubMed id:
|
|
|
|

|
| |
|
Atomic description of an enzyme reaction dominated by proton tunneling.
|
|
L.Masgrau,
A.Roujeinikova,
L.O.Johannissen,
P.Hothi,
J.Basran,
K.E.Ranaghan,
A.J.Mulholland,
M.J.Sutcliffe,
N.S.Scrutton,
D.Leys.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
We present an atomic-level description of the reaction chemistry of an
enzyme-catalyzed reaction dominated by proton tunneling. By solving structures
of reaction intermediates at near-atomic resolution, we have identified the
reaction pathway for tryptamine oxidation by aromatic amine dehydrogenase.
Combining experiment and computer simulation, we show proton transfer occurs
predominantly to oxygen O2 of Asp(128)beta in a reaction dominated by tunneling
over approximately 0.6 angstroms. The role of long-range coupled motions in
promoting tunneling is controversial. We show that, in this enzyme system,
tunneling is promoted by a short-range motion modulating proton-acceptor
distance and no long-range coupled motion is required.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Fig. 1. Schematic overview of the AADH reaction with
tryptamine. The AADH structure is represented in cartoon form
with the  subunit in blue,
the ß subunit in yellow, and the TTQ cofactor in magenta
spheres. The reductive half-reaction with tryptamine is depicted
in detail in Fig. 2. The oxidative half-reaction consists of two
consecutive long-range electron transfer events to the
single-electron carrier azurin. An azurin from Pseudomonas
putida (PDB code 1NWO) is depicted in green cartoon with the
copper atom as an orange sphere. subunit in blue,
the ß subunit in yellow, and the TTQ cofactor in magenta
spheres. The reductive half-reaction with tryptamine is depicted
in detail in Fig. 2. The oxidative half-reaction consists of two
consecutive long-range electron transfer events to the
single-electron carrier azurin. An azurin from Pseudomonas
putida (PDB code 1NWO) is depicted in green cartoon with the
copper atom as an orange sphere.
|
 |
Figure 2.
Fig. 2. Reductive half-reaction of AADH with tryptamine. (A)
Schematic overview of the proposed reaction mechanism.
Intermediates are numbered by roman numerals; for clarity only
key atoms are represented from intermediate II onward, whereas
TTQ atoms C6 and C7 and Asp128ß O1 and O2 are labeled for
intermediate I. Atoms derived from the substrate are depicted in
magenta; those derived from water, in blue. All other
enzyme-derived atoms are depicted in black. (B) AADH active site
structure for the ligand-free protein, represented by
atom-colored sticks: green carbons, TTQ cofactor; other carbons,
yellow. The position occupied by the substrate in complex with
the reduced AADH is depicted in dark gray, and the positions in
intermediate V of substrate Leu179  and TTQ are in
light gray; putative hydrogen bonds are shown as green dashed
lines. (C) Active site and SigmaA weighted F[o]F[c] and
2F[o]F[c]-electron density of intermediates in the reductive
half-reaction with tryptamine. Atoms are colored according to
atom type, with substrate-derived carbon atoms in magenta and
protein-derived carbon atoms in yellow. For clarity, only
substrate- or inhibitor-derived atoms are shown, in addition to
the side chain atoms for residues Asp128ß and
Thr172ß and part of the TTQ cofactor. The 2F[o]F[c]
density for these enzyme-derived atoms or water molecules is
shown as a blue mesh, whereas F[o]F[c] density for the
substrate- or inhibitor-derived atoms is depicted at 4 and TTQ are in
light gray; putative hydrogen bonds are shown as green dashed
lines. (C) Active site and SigmaA weighted F[o]F[c] and
2F[o]F[c]-electron density of intermediates in the reductive
half-reaction with tryptamine. Atoms are colored according to
atom type, with substrate-derived carbon atoms in magenta and
protein-derived carbon atoms in yellow. For clarity, only
substrate- or inhibitor-derived atoms are shown, in addition to
the side chain atoms for residues Asp128ß and
Thr172ß and part of the TTQ cofactor. The 2F[o]F[c]
density for these enzyme-derived atoms or water molecules is
shown as a blue mesh, whereas F[o]F[c] density for the
substrate- or inhibitor-derived atoms is depicted at 4  and shown as a
magenta mesh. Intermediates are labeled by roman numerals
corresponding to (A). The covalent phenylhydrazine-AADH complex
is used as a model for intermediate III. For intermediate V,
electron density for the P2[1] crystal form is shown, whereas
intermediate VII is depicted by electron density for the F222
crystal form. and shown as a
magenta mesh. Intermediates are labeled by roman numerals
corresponding to (A). The covalent phenylhydrazine-AADH complex
is used as a model for intermediate III. For intermediate V,
electron density for the P2[1] crystal form is shown, whereas
intermediate VII is depicted by electron density for the F222
crystal form.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the AAAs:
Science
(2006,
312,
237-241)
copyright 2006.
|
|
| |
Figures were
selected
by the author.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
X.Z.Li,
B.Walker,
and
A.Michaelides
(2011).
Quantum nature of the hydrogen bond.
|
| |
Proc Natl Acad Sci U S A,
108,
6369-6373.
|
 |
|
|
|
|
 |
B.R.Menon,
P.A.Davison,
C.N.Hunter,
N.S.Scrutton,
and
D.J.Heyes
(2010).
Mutagenesis alters the catalytic mechanism of the light-driven enzyme protochlorophyllide oxidoreductase.
|
| |
J Biol Chem,
285,
2113-2119.
|
 |
|
|
|
|
 |
D.Murdock,
L.A.Burns,
and
P.H.Vaccaro
(2010).
Vibrational specificity of proton-transfer dynamics in ground-state tropolone.
|
| |
Phys Chem Chem Phys,
12,
8285-8299.
|
 |
|
|
|
|
 |
D.Roston,
and
A.Kohen
(2010).
Elusive transition state of alcohol dehydrogenase unveiled.
|
| |
Proc Natl Acad Sci U S A,
107,
9572-9577.
|
 |
|
|
|
|
 |
H.Dong,
S.Qin,
and
H.X.Zhou
(2010).
Effects of macromolecular crowding on protein conformational changes.
|
| |
PLoS Comput Biol,
6,
e1000833.
|
 |
|
|
|
|
 |
J.Pang,
N.S.Scrutton,
S.P.de Visser,
and
M.J.Sutcliffe
(2010).
New insights into the multi-step reaction pathway of the reductive half-reaction catalysed by aromatic amine dehydrogenase: a QM/MM study.
|
| |
Chem Commun (Camb),
46,
3104-3106.
|
 |
|
|
|
|
 |
M.Yoon,
H.Song,
K.Håkansson,
and
E.N.Marsh
(2010).
Hydrogen tunneling in adenosylcobalamin-dependent glutamate mutase: evidence from intrinsic kinetic isotope effects measured by intramolecular competition.
|
| |
Biochemistry,
49,
3168-3173.
|
 |
|
|
|
|
 |
R.A.Mata
(2010).
Application of high level wavefunction methods in quantum mechanics/molecular mechanics hybrid schemes.
|
| |
Phys Chem Chem Phys,
12,
5041-5052.
|
 |
|
|
|
|
 |
R.Lonsdale,
K.E.Ranaghan,
and
A.J.Mulholland
(2010).
Computational enzymology.
|
| |
Chem Commun (Camb),
46,
2354-2372.
|
 |
|
|
|
|
 |
S.C.Kamerlin,
and
A.Warshel
(2010).
At the dawn of the 21st century: Is dynamics the missing link for understanding enzyme catalysis?
|
| |
Proteins,
78,
1339-1375.
|
 |
|
|
|
|
 |
S.Hay,
L.O.Johannissen,
M.J.Sutcliffe,
and
N.S.Scrutton
(2010).
Barrier compression and its contribution to both classical and quantum mechanical aspects of enzyme catalysis.
|
| |
Biophys J,
98,
121-128.
|
 |
|
|
|
|
 |
T.Zimmermann,
and
J.Vaníček
(2010).
Three applications of path integrals: equilibrium and kinetic isotope effects, and the temperature dependence of the rate constant of the [1,5] sigmatropic hydrogen shift in (Z)-1,3-pentadiene.
|
| |
J Mol Model,
16,
1779-1787.
|
 |
|
|
|
|
 |
A.Yahashiri,
G.Nimrod,
N.Ben-Tal,
E.E.Howell,
and
A.Kohen
(2009).
The effect of electrostatic shielding on H tunneling in R67 dihydrofolate reductase.
|
| |
Chembiochem,
10,
2620-2623.
|
 |
|
|
|
|
 |
E.F.Garman,
and
C.Nave
(2009).
Radiation damage in protein crystals examined under various conditions by different methods.
|
| |
J Synchrotron Radiat,
16,
129-132.
|
 |
|
|
|
|
 |
J.N.Bandaria,
C.M.Cheatum,
and
A.Kohen
(2009).
Examination of enzymatic H-tunneling through kinetics and dynamics.
|
| |
J Am Chem Soc,
131,
10151-10155.
|
 |
|
|
|
|
 |
M.Arndt,
T.Juffmann,
and
V.Vedral
(2009).
Quantum physics meets biology.
|
| |
HFSP J,
3,
386-400.
|
 |
|
|
|
|
 |
R.Baron,
C.Riley,
P.Chenprakhon,
K.Thotsaporn,
R.T.Winter,
A.Alfieri,
F.Forneris,
W.J.van Berkel,
P.Chaiyen,
M.W.Fraaije,
A.Mattevi,
and
J.A.McCammon
(2009).
Multiple pathways guide oxygen diffusion into flavoenzyme active sites.
|
| |
Proc Natl Acad Sci U S A,
106,
10603-10608.
|
 |
|
|
|
|
 |
S.Hay,
C.R.Pudney,
and
N.S.Scrutton
(2009).
Structural and mechanistic aspects of flavoproteins: probes of hydrogen tunnelling.
|
| |
FEBS J,
276,
3930-3941.
|
 |
|
|
|
|
 |
A.J.Mulholland
(2008).
Introduction. Biomolecular simulation.
|
| |
J R Soc Interface,
5,
S169-S172.
|
 |
|
|
|
|
 |
A.Yahashiri,
E.E.Howell,
and
A.Kohen
(2008).
Tuning of the H-transfer coordinate in primitive versus well-evolved enzymes.
|
| |
Chemphyschem,
9,
980-982.
|
 |
|
|
|
|
 |
C.J.Woods,
F.R.Manby,
and
A.J.Mulholland
(2008).
An efficient method for the calculation of quantum mechanics/molecular mechanics free energies.
|
| |
J Chem Phys,
128,
014109.
|
 |
|
|
|
|
 |
E.J.Loveridge,
R.M.Evans,
and
R.K.Allemann
(2008).
Solvent effects on environmentally coupled hydrogen tunnelling during catalysis by dihydrofolate reductase from Thermotoga maritima.
|
| |
Chemistry,
14,
10782-10788.
|
 |
|
|
|
|
 |
G.G.Dodson,
D.P.Lane,
and
C.S.Verma
(2008).
Molecular simulations of protein dynamics: new windows on mechanisms in biology.
|
| |
EMBO Rep,
9,
144-150.
|
 |
|
|
|
|
 |
J.N.Bandaria,
S.Dutta,
S.E.Hill,
A.Kohen,
and
C.M.Cheatum
(2008).
Fast enzyme dynamics at the active site of formate dehydrogenase.
|
| |
J Am Chem Soc,
130,
22-23.
|
 |
|
|
|
|
 |
L.O.Johannissen,
N.S.Scrutton,
and
M.J.Sutcliffe
(2008).
The enzyme aromatic amine dehydrogenase induces a substrate conformation crucial for promoting vibration that significantly reduces the effective potential energy barrier to proton transfer.
|
| |
J R Soc Interface,
5,
S225-S232.
|
 |
|
|
|
|
 |
L.Swint-Kruse,
and
H.F.Fisher
(2008).
Enzymatic reaction sequences as coupled multiple traces on a multidimensional landscape.
|
| |
Trends Biochem Sci,
33,
104-112.
|
 |
|
|
|
|
 |
M.Koutmos,
R.Pejchal,
T.M.Bomer,
R.G.Matthews,
J.L.Smith,
and
M.L.Ludwig
(2008).
Metal active site elasticity linked to activation of homocysteine in methionine synthases.
|
| |
Proc Natl Acad Sci U S A,
105,
3286-3291.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.W.van der Kamp,
and
A.J.Mulholland
(2008).
Computational enzymology: insight into biological catalysts from modelling.
|
| |
Nat Prod Rep,
25,
1001-1014.
|
 |
|
|
|
|
 |
O.A.Sytina,
D.J.Heyes,
C.N.Hunter,
M.T.Alexandre,
I.H.van Stokkum,
R.van Grondelle,
and
M.L.Groot
(2008).
Conformational changes in an ultrafast light-driven enzyme determine catalytic activity.
|
| |
Nature,
456,
1001-1004.
|
 |
|
|
|
|
 |
P.Hothi,
S.Hay,
A.Roujeinikova,
M.J.Sutcliffe,
M.Lee,
D.Leys,
P.M.Cullis,
and
N.S.Scrutton
(2008).
Driving force analysis of proton tunnelling across a reactivity series for an enzyme-substrate complex.
|
| |
Chembiochem,
9,
2839-2845.
|
 |
|
|
|
|
 |
R.H.Steele
(2008).
Harmonic oscillators: the quantization of simple systems in the old quantum theory and their functional roles in biology.
|
| |
Mol Cell Biochem,
310,
19-42.
|
 |
|
|
|
|
 |
S.Hay,
C.R.Pudney,
M.J.Sutcliffe,
and
N.S.Scrutton
(2008).
Solvent as a probe of active site motion and chemistry during the hydrogen tunnelling reaction in morphinone reductase.
|
| |
Chemphyschem,
9,
1875-1881.
|
 |
|
|
|
|
 |
S.Hay,
J.Pang,
P.J.Monaghan,
X.Wang,
R.M.Evans,
M.J.Sutcliffe,
R.K.Allemann,
and
N.S.Scrutton
(2008).
Secondary kinetic isotope effects as probes of environmentally-coupled enzymatic hydrogen tunneling reactions.
|
| |
Chemphyschem,
9,
1536-1539.
|
 |
|
|
|
|
 |
S.Hay,
and
N.S.Scrutton
(2008).
H-transfers in Photosystem II: what can we learn from recent lessons in the enzyme community?
|
| |
Photosynth Res,
98,
169-177.
|
 |
|
|
|
|
 |
S.Saen-Oon,
M.Ghanem,
V.L.Schramm,
and
S.D.Schwartz
(2008).
Remote mutations and active site dynamics correlate with catalytic properties of purine nucleoside phosphorylase.
|
| |
Biophys J,
94,
4078-4088.
|
 |
|
|
|
|
 |
U.Pentikäinen,
O.T.Pentikäinen,
and
A.J.Mulholland
(2008).
Cooperative symmetric to asymmetric conformational transition of the apo-form of scavenger decapping enzyme revealed by simulations.
|
| |
Proteins,
70,
498-508.
|
 |
|
|
|
|
 |
A.J.Mulholland
(2007).
Chemical accuracy in QM/MM calculations on enzyme-catalysed reactions.
|
| |
Chem Cent J,
1,
19.
|
 |
|
|
|
|
 |
A.Lodola,
M.Mor,
J.Zurek,
G.Tarzia,
D.Piomelli,
J.N.Harvey,
and
A.J.Mulholland
(2007).
Conformational effects in enzyme catalysis: reaction via a high energy conformation in fatty acid amide hydrolase.
|
| |
Biophys J,
92,
L20-L22.
|
 |
|
|
|
|
 |
D.Zhong
(2007).
Ultrafast catalytic processes in enzymes.
|
| |
Curr Opin Chem Biol,
11,
174-181.
|
 |
|
|
|
|
 |
J.N.Harvey
(2007).
Understanding the kinetics of spin-forbidden chemical reactions.
|
| |
Phys Chem Chem Phys,
9,
331-343.
|
 |
|
|
|
|
 |
K.E.Ranaghan,
L.Masgrau,
N.S.Scrutton,
M.J.Sutcliffe,
and
A.J.Mulholland
(2007).
Analysis of classical and quantum paths for deprotonation of methylamine by methylamine dehydrogenase.
|
| |
Chemphyschem,
8,
1816-1835.
|
 |
|
|
|
|
 |
M.W.van der Kamp,
F.Perruccio,
and
A.J.Mulholland
(2007).
Substrate polarization in enzyme catalysis: QM/MM analysis of the effect of oxaloacetate polarization on acetyl-CoA enolization in citrate synthase.
|
| |
Proteins,
69,
521-535.
|
 |
|
|
|
|
 |
S.Hay,
M.J.Sutcliffe,
and
N.S.Scrutton
(2007).
Promoting motions in enzyme catalysis probed by pressure studies of kinetic isotope effects.
|
| |
Proc Natl Acad Sci U S A,
104,
507-512.
|
 |
|
|
|
|
 |
Y.T.Kao,
C.Saxena,
L.Wang,
A.Sancar,
and
D.Zhong
(2007).
Femtochemistry in enzyme catalysis: DNA photolyase.
|
| |
Cell Biochem Biophys,
48,
32-44.
|
 |
|
|
|
|
 |
F.Claeyssens,
J.N.Harvey,
F.R.Manby,
R.A.Mata,
A.J.Mulholland,
K.E.Ranaghan,
M.Schütz,
S.Thiel,
W.Thiel,
and
H.J.Werner
(2006).
High-accuracy computation of reaction barriers in enzymes.
|
| |
Angew Chem Int Ed Engl,
45,
6856-6859.
|
 |
|
|
|
|
 |
J.Zurek,
N.Foloppe,
J.N.Harvey,
and
A.J.Mulholland
(2006).
Mechanisms of reaction in cytochrome P450: Hydroxylation of camphor in P450cam.
|
| |
Org Biomol Chem,
4,
3931-3937.
|
 |
|
|
|
|
 |
M.J.Sutcliffe,
L.Masgrau,
A.Roujeinikova,
L.O.Johannissen,
P.Hothi,
J.Basran,
K.E.Ranaghan,
A.J.Mulholland,
D.Leys,
and
N.S.Scrutton
(2006).
Hydrogen tunnelling in enzyme-catalysed H-transfer reactions: flavoprotein and quinoprotein systems.
|
| |
Philos Trans R Soc Lond B Biol Sci,
361,
1375-1386.
|
 |
|
|
|
|
 |
M.J.Sutcliffe,
and
N.S.Scrutton
(2006).
Computational studies of enzyme mechanism: linking theory with experiment in the analysis of enzymic H-tunnelling.
|
| |
Phys Chem Chem Phys,
8,
4510-4516.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

