 |
PDBsum entry 2a8t

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Translation,hydrolase
|
PDB id
|
|

|

|
2a8t
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.3.6.1.62
- 5'-(N(7)-methylguanosine 5'-triphospho)-[mRNA] hydrolase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a 5'-end (N7-methyl 5'-triphosphoguanosine)-ribonucleoside in mRNA + H2O = N(7)-methyl-GDP + a 5'-end phospho-ribonucleoside in mRNA + 2 H+
|
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.3.6.1.64
- inosine diphosphate phosphatase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
IDP + H2O = IMP + phosphate + H+
|
|
2.
|
dIDP + H2O = dIMP + phosphate + H+
|
|
 |
 |
 |
 |
 |
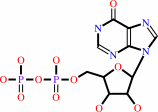 IDP
IDP
|
+
|
H2O
Bound ligand (Het Group name = )
matches with 84.38% similarity
|
=
|
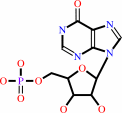 IMP
IMP
|
+
|
 phosphate
phosphate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
dIDP
Bound ligand (Het Group name = )
matches with 81.25% similarity
|
+
|
H2O
|
=
|
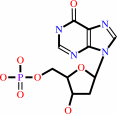 dIMP
dIMP
|
+
|
 phosphate
phosphate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Structure
14:331-343
(2006)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structures of U8 snoRNA decapping nudix hydrolase, X29, and its metal and cap complexes.
|
|
J.N.Scarsdale,
B.A.Peculis,
H.T.Wright.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
X29, a 25 kDa Nudix hydrolase from Xenopus laevis that cleaves 5' caps from U8
snoRNA, crystallizes as a homodimeric apoenzyme. Manganese binds crystals of
apo-X29 to form holo-X29 only in the presence of nucleot(s)ide. Structural
changes in X29 on nucleo-t(s)ide-assisted Mn(+2) uptake account for the observed
cooperativity of metal binding. Structures of X29 with GTP or m7GpppA show a
different mode of ligand binding from that of other cap binding proteins and
suggest a possible three- or four-metal Nudix reaction mechanism. The X29 dimer
has no known RNA binding motif, but its striking surface dipolarity and unique
structural features create a plausible RNA binding channel on the positive face
of the protein. Because U8 snoRNP is essential for accumulation of mature 5.8S
and 28S rRNA in vertebrate ribosome biogenesis, and cap structures are required
for U8 stability in vivo, X29 could profoundly influence this fundamental
cellular pathway.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |
Figure 3.
Figure 3. Surface Charge Distribution of Opposite Faces of
X29
Blue indicates regions of positive surface charge and
red indicates negative, contoured at ± 5kt cutoff, respectively.
Overlaid ribbon in (C) shows extensions (black arrow pointers)
that define the two sides of the inferred RNA binding channel,
which is indicated by yellow arrows. Active sites of each
monomer are occupied by m7GpppA (gold and green ball-and-stick).
Views are down the dyad axis.
|
 |
|
|

|
| |
The above figure is
reprinted
by permission from Cell Press:
Structure
(2006,
14,
331-343)
copyright 2006.
|
|
| |
Figure was
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
G.Lu,
J.Zhang,
Y.Li,
Z.Li,
N.Zhang,
X.Xu,
T.Wang,
Z.Guan,
G.F.Gao,
and
J.Yan
(2011).
hNUDT16: a universal decapping enzyme for small nucleolar RNA and cytoplasmic mRNA.
|
| |
Protein Cell,
2,
64-73.
|
 |
|
|
|
|
 |
M.F.Soulière,
J.P.Perreault,
and
M.Bisaillon
(2010).
Insights into the molecular determinants involved in cap recognition by the vaccinia virus D10 decapping enzyme.
|
| |
Nucleic Acids Res,
38,
7599-7610.
|
 |
|
|
|
|
 |
T.Nakamura,
S.Meshitsuka,
S.Kitagawa,
N.Abe,
J.Yamada,
T.Ishino,
H.Nakano,
T.Tsuzuki,
T.Doi,
Y.Kobayashi,
S.Fujii,
M.Sekiguchi,
and
Y.Yamagata
(2010).
Structural and dynamic features of the MutT protein in the recognition of nucleotides with the mutagenic 8-oxoguanine base.
|
| |
J Biol Chem,
285,
444-452.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.G.Thorsell,
C.Persson,
S.Gräslund,
M.Hammarström,
R.D.Busam,
and
B.M.Hallberg
(2009).
Crystal structure of human diphosphoinositol phosphatase 1.
|
| |
Proteins,
77,
242-246.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.A.Messing,
S.B.Gabelli,
Q.Liu,
H.Celesnik,
J.G.Belasco,
S.A.Piñeiro,
and
L.M.Amzel
(2009).
Structure and biological function of the RNA pyrophosphohydrolase BdRppH from Bdellovibrio bacteriovorus.
|
| |
Structure,
17,
472-481.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Bail,
and
M.Kiledjian
(2009).
Tri- to be mono- for bacterial mRNA decay.
|
| |
Structure,
17,
317-319.
|
 |
|
|
|
|
 |
Y.Li,
E.S.Ho,
S.I.Gunderson,
and
M.Kiledjian
(2009).
Mutational analysis of a Dcp2-binding element reveals general enhancement of decapping by 5'-end stem-loop structures.
|
| |
Nucleic Acids Res,
37,
2227-2237.
|
 |
|
|
|
|
 |
J.Zhang,
F.Gao,
Q.Zhang,
Q.Chen,
J.Qi,
and
J.Yan
(2008).
Crystallization and crystallographic analysis of human NUDT16.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
64,
639-640.
|
 |
|
|
|
|
 |
M.J.Taylor,
and
B.A.Peculis
(2008).
Evolutionary conservation supports ancient origin for Nudt16, a nuclear-localized, RNA-binding, RNA-decapping enzyme.
|
| |
Nucleic Acids Res,
36,
6021-6034.
|
 |
|
|
|
|
 |
B.A.Peculis,
K.Reynolds,
and
M.Cleland
(2007).
Metal determines efficiency and substrate specificity of the nuclear NUDIX decapping proteins X29 and H29K (Nudt16).
|
| |
J Biol Chem,
282,
24792-24805.
|
 |
|
|
|
|
 |
V.Shen,
and
M.Kiledjian
(2006).
Decapper comes into focus.
|
| |
Structure,
14,
171-172.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

