 |
PDBsum entry 1xn0

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase
|
 |
|
Title:
|
 |
Catalytic domain of human phosphodiesterase 4b in complex with (r,s)- rolipram
|
|
Structure:
|
 |
Camp-specific 3',5'-cyclic phosphodiesterase 4b. Chain: a, b. Fragment: catalytic domain of human phosphodiesterase 4b. Synonym: dpde4, pde32. Engineered: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Gene: pde4b. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
2.31Å
|
R-factor:
|
0.208
|
R-free:
|
0.250
|
|
|
Authors:
|
 |
G.L.Card,B.P.England,Y.Suzuki,D.Fong,B.Powell,B.Lee,C.Luu, M.Tabrizizad,S.Gillette,P.N.Ibrahim,D.R.Artis,G.Bollag,M.V.Milburn, S.-H.Kim,J.Schlessinger,K.Y.J.Zhang
|
Key ref:

|
 |
G.L.Card
et al.
(2004).
Structural basis for the activity of drugs that inhibit phosphodiesterases.
Structure,
12,
2233-2247.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
04-Oct-04
|
Release date:
|
14-Dec-04
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q07343
(PDE4B_HUMAN) -
3',5'-cyclic-AMP phosphodiesterase 4B from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
736 a.a.
323 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 3 residue positions (black
crosses)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.1.4.53
- 3',5'-cyclic-AMP phosphodiesterase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
3',5'-cyclic AMP + H2O = AMP + H+
|
 |
 |
 |
 |
 |
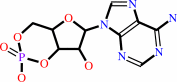 3',5'-cyclic AMP
3',5'-cyclic AMP
|
+
|
H2O
|
=
|
 AMP
AMP
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Structure
12:2233-2247
(2004)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural basis for the activity of drugs that inhibit phosphodiesterases.
|
|
G.L.Card,
B.P.England,
Y.Suzuki,
D.Fong,
B.Powell,
B.Lee,
C.Luu,
M.Tabrizizad,
S.Gillette,
P.N.Ibrahim,
D.R.Artis,
G.Bollag,
M.V.Milburn,
S.H.Kim,
J.Schlessinger,
K.Y.Zhang.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Phosphodiesterases (PDEs) comprise a large family of enzymes that catalyze the
hydrolysis of cAMP or cGMP and are implicated in various diseases. We describe
the high-resolution crystal structures of the catalytic domains of PDE4B, PDE4D,
and PDE5A with ten different inhibitors, including the drug candidates
cilomilast and roflumilast, for respiratory diseases. These cocrystal structures
reveal a common scheme of inhibitor binding to the PDEs: (i) a hydrophobic clamp
formed by highly conserved hydrophobic residues that sandwich the inhibitor in
the active site; (ii) hydrogen bonding to an invariant glutamine that controls
the orientation of inhibitor binding. A scaffold can be readily identified for
any given inhibitor based on the formation of these two types of conserved
interactions. These structural insights will enable the design of
isoform-selective inhibitors with improved binding affinity and should
facilitate the discovery of more potent and selective PDE inhibitors for the
treatment of a variety of diseases.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |
Figure 1.
Figure 1. Classification of the Active Site of PDEs(A) The
active site of PDEs is divided into three pockets: the metal
binding pocket (M) shown in blue, the purine-selective glutamine
and hydrophobic clamp pocket (Q) shown in red (which is further
divided into Q[1] and Q[2] subpockets), and the solvent-filled
side pocket (S) shown in green. This color coding of the active
site pocket is mapped on the surface of PDE4B in complex with
cilomilast, which is shown as a stick model bound at the active
site. The cocrystal structure of PDE4B in complex with
cilomilast has also been used to display the surfaces in
(B)-(D).(B) Same as (A), but a view of the PDE active site
looking toward the S pocket. This view is a clockwise rotation
of about 90° along the length of cilomilast from the view in
Figure 1A. The subpockets that subdivide the Q pocket are also
labeled: Q[1] is the small subpocket, and Q[2] is the large
subpocket.(C) Same as (A), but a view of the PDE active site
looking away from the S pocket. This view is a counterclockwise
rotation of about 90° along the length of cilomilast from the
view in Figure 1A. All the subpockets are labeled.(D) Residues
lining the three active site pockets. The active site surface is
semitransparent to reveal residues that make up the active site.
The absolutely conserved residues in all PDEs are colored blue.
Residues conserved in both cAMP- and cGMP-specific PDEs are
colored green. The other variable residues are colored red.
|
 |
|
|

|
| |
The above figure is
reprinted
by permission from Cell Press:
Structure
(2004,
12,
2233-2247)
copyright 2004.
|
|
| |
Figure was
selected
by the author.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
R.W.Allcock,
H.Blakli,
Z.Jiang,
K.A.Johnston,
K.M.Morgan,
G.M.Rosair,
K.Iwase,
Y.Kohno,
and
D.R.Adams
(2011).
Phosphodiesterase inhibitors. Part 1: Synthesis and structure-activity relationships of pyrazolopyridine-pyridazinone PDE inhibitors developed from ibudilast.
|
| |
Bioorg Med Chem Lett,
21,
3307-3312.
|
 |
|
|
|
|
 |
A.B.Burgin,
O.T.Magnusson,
J.Singh,
P.Witte,
B.L.Staker,
J.M.Bjornsson,
M.Thorsteinsdottir,
S.Hrafnsdottir,
T.Hagen,
A.S.Kiselyov,
L.J.Stewart,
and
M.E.Gurney
(2010).
Design of phosphodiesterase 4D (PDE4D) allosteric modulators for enhancing cognition with improved safety.
|
| |
Nat Biotechnol,
28,
63-70.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
K.Murata,
N.Nagata,
I.Nakanishi,
and
K.Kitaura
(2010).
SDOVS: a solvent dipole ordering-based method for virtual screening.
|
| |
J Comput Chem,
31,
2714-2722.
|
 |
|
|
|
|
 |
M.A.Giembycz,
and
S.K.Field
(2010).
Roflumilast: first phosphodiesterase 4 inhibitor approved for treatment of COPD.
|
| |
Drug Des Devel Ther,
4,
147-158.
|
 |
|
|
|
|
 |
P.V.Mazin,
M.S.Gelfand,
A.A.Mironov,
A.B.Rakhmaninova,
A.R.Rubinov,
R.B.Russell,
and
O.V.Kalinina
(2010).
An automated stochastic approach to the identification of the protein specificity determinants and functional subfamilies.
|
| |
Algorithms Mol Biol,
5,
29.
|
 |
|
|
|
|
 |
S.King-Keller,
M.Li,
A.Smith,
S.Zheng,
G.Kaur,
X.Yang,
B.Wang,
and
R.Docampo
(2010).
Chemical validation of phosphodiesterase C as a chemotherapeutic target in Trypanosoma cruzi, the etiological agent of Chagas' disease.
|
| |
Antimicrob Agents Chemother,
54,
3738-3745.
|
 |
|
|
|
|
 |
A.P.Skoumbourdis,
C.A.Leclair,
E.Stefan,
A.G.Turjanski,
W.Maguire,
S.A.Titus,
R.Huang,
D.S.Auld,
J.Inglese,
C.P.Austin,
S.W.Michnick,
M.Xia,
and
C.J.Thomas
(2009).
Exploration and optimization of substituted triazolothiadiazines and triazolopyridazines as PDE4 inhibitors.
|
| |
Bioorg Med Chem Lett,
19,
3686-3692.
|
 |
|
|
|
|
 |
B.Barren,
L.Gakhar,
H.Muradov,
K.K.Boyd,
S.Ramaswamy,
and
N.O.Artemyev
(2009).
Structural basis of phosphodiesterase 6 inhibition by the C-terminal region of the gamma-subunit.
|
| |
EMBO J,
28,
3613-3622.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Y.Sato,
Y.Hashiguchi,
and
M.Nishida
(2009).
Evolution of multiple phosphodiesterase isoforms in stickleback involved in cAMP signal transduction pathway.
|
| |
BMC Syst Biol,
3,
23.
|
 |
|
|
|
|
 |
C.C.Heikaus,
J.R.Stout,
M.R.Sekharan,
C.M.Eakin,
P.Rajagopal,
P.S.Brzovic,
J.A.Beavo,
and
R.E.Klevit
(2008).
Solution structure of the cGMP binding GAF domain from phosphodiesterase 5: insights into nucleotide specificity, dimerization, and cGMP-dependent conformational change.
|
| |
J Biol Chem,
283,
22749-22759.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
D.Spina
(2008).
PDE4 inhibitors: current status.
|
| |
Br J Pharmacol,
155,
308-315.
|
 |
|
|
|
|
 |
G.Chen,
H.Wang,
H.Robinson,
J.Cai,
Y.Wan,
and
H.Ke
(2008).
An insight into the pharmacophores of phosphodiesterase-5 inhibitors from synthetic and crystal structural studies.
|
| |
Biochem Pharmacol,
75,
1717-1728.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
H.Wang,
M.Ye,
H.Robinson,
S.H.Francis,
and
H.Ke
(2008).
Conformational variations of both phosphodiesterase-5 and inhibitors provide the structural basis for the physiological effects of vardenafil and sildenafil.
|
| |
Mol Pharmacol,
73,
104-110.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.K.Field
(2008).
Roflumilast: an oral, once-daily selective PDE-4 inhibitor for the management of COPD and asthma.
|
| |
Expert Opin Investig Drugs,
17,
811-818.
|
 |
|
|
|
|
 |
X.J.Zhang,
K.B.Cahill,
A.Elfenbein,
V.Y.Arshavsky,
and
R.H.Cote
(2008).
Direct Allosteric Regulation between the GAF Domain and Catalytic Domain of Photoreceptor Phosphodiesterase PDE6.
|
| |
J Biol Chem,
283,
29699-29705.
|
 |
|
|
|
|
 |
Y.Xiong,
H.T.Lu,
and
C.G.Zhan
(2008).
Dynamic structures of phosphodiesterase-5 active site by combined molecular dynamics simulations and hybrid quantum mechanical/molecular mechanical calculations.
|
| |
J Comput Chem,
29,
1259-1267.
|
 |
|
|
|
|
 |
M.Conti,
and
J.Beavo
(2007).
Biochemistry and physiology of cyclic nucleotide phosphodiesterases: essential components in cyclic nucleotide signaling.
|
| |
Annu Rev Biochem,
76,
481-511.
|
 |
|
|
|
|
 |
W.M.Brown
(2007).
Treating COPD with PDE 4 inhibitors.
|
| |
Int J Chron Obstruct Pulmon Dis,
2,
517-533.
|
 |
|
|
|
|
 |
A.Ghavami,
W.D.Hirst,
and
T.J.Novak
(2006).
Selective phosphodiesterase (PDE)-4 inhibitors: a novel approach to treating memory deficit?
|
| |
Drugs R D,
7,
63-71.
|
 |
|
|
|
|
 |
A.Johner,
S.Kunz,
M.Linder,
Y.Shakur,
and
T.Seebeck
(2006).
Cyclic nucleotide specific phosphodiesterases of Leishmania major.
|
| |
BMC Microbiol,
6,
25.
|
 |
|
|
|
|
 |
D.Wang,
and
X.Cui
(2006).
Evaluation of PDE4 inhibition for COPD.
|
| |
Int J Chron Obstruct Pulmon Dis,
1,
373-379.
|
 |
|
|
|
|
 |
F.G.Oliveira,
C.M.Sant'Anna,
E.R.Caffarena,
L.E.Dardenne,
and
E.J.Barreiro
(2006).
Molecular docking study and development of an empirical binding free energy model for phosphodiesterase 4 inhibitors.
|
| |
Bioorg Med Chem,
14,
6001-6011.
|
 |
|
|
|
|
 |
F.S.Menniti,
W.S.Faraci,
and
C.J.Schmidt
(2006).
Phosphodiesterases in the CNS: targets for drug development.
|
| |
Nat Rev Drug Discov,
5,
660-670.
|
 |
|
|
|
|
 |
H.Park,
J.Lee,
and
S.Lee
(2006).
Critical assessment of the automated AutoDock as a new docking tool for virtual screening.
|
| |
Proteins,
65,
549-554.
|
 |
|
|
|
|
 |
Q.Huai,
Y.Sun,
H.Wang,
D.Macdonald,
R.Aspiotis,
H.Robinson,
Z.Huang,
and
H.Ke
(2006).
Enantiomer discrimination illustrated by the high resolution crystal structures of type 4 phosphodiesterase.
|
| |
J Med Chem,
49,
1867-1873.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Y.Xiong,
H.T.Lu,
Y.Li,
G.F.Yang,
and
C.G.Zhan
(2006).
Characterization of a catalytic ligand bridging metal ions in phosphodiesterases 4 and 5 by molecular dynamics simulations and hybrid quantum mechanical/molecular mechanical calculations.
|
| |
Biophys J,
91,
1858-1867.
|
 |
|
|
|
|
 |
G.L.Card,
L.Blasdel,
B.P.England,
C.Zhang,
Y.Suzuki,
S.Gillette,
D.Fong,
P.N.Ibrahim,
D.R.Artis,
G.Bollag,
M.V.Milburn,
S.H.Kim,
J.Schlessinger,
and
K.Y.Zhang
(2005).
A family of phosphodiesterase inhibitors discovered by cocrystallography and scaffold-based drug design.
|
| |
Nat Biotechnol,
23,
201-207.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
H.Wang,
Y.Liu,
Y.Chen,
H.Robinson,
and
H.Ke
(2005).
Multiple elements jointly determine inhibitor selectivity of cyclic nucleotide phosphodiesterases 4 and 7.
|
| |
J Biol Chem,
280,
30949-30955.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
K.Y.Zhang,
P.N.Ibrahim,
S.Gillette,
and
G.Bollag
(2005).
Phosphodiesterase-4 as a potential drug target.
|
| |
Expert Opin Ther Targets,
9,
1283-1305.
|
 |
|
|
|
|
 |
M.D.Houslay,
P.Schafer,
and
K.Y.Zhang
(2005).
Keynote review: phosphodiesterase-4 as a therapeutic target.
|
| |
Drug Discov Today,
10,
1503-1519.
|
 |
|
|
|
|
 |
S.Kunz,
M.Oberholzer,
and
T.Seebeck
(2005).
A FYVE-containing unusual cyclic nucleotide phosphodiesterase from Trypanosoma cruzi.
|
| |
FEBS J,
272,
6412-6422.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

