 |
PDBsum entry 1x9e

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.3.3.10
- hydroxymethylglutaryl-CoA synthase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Mevalonate Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
acetoacetyl-CoA + acetyl-CoA + H2O = (3S)-3-hydroxy-3-methylglutaryl-CoA + CoA + H+
|
 |
 |
 |
 |
 |
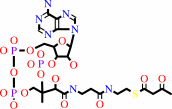 acetoacetyl-CoA
acetoacetyl-CoA
|
+
|
 acetyl-CoA
acetyl-CoA
|
+
|
H2O
|
=
|
(3S)-3-hydroxy-3-methylglutaryl-CoA
|
+
|
 CoA
CoA
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
44:14256-14267
(2005)
|
|
PubMed id:
|
|
|
|

|
| |
|
X-ray crystal structures of HMG-CoA synthase from Enterococcus faecalis and a complex with its second substrate/inhibitor acetoacetyl-CoA.
|
|
C.N.Steussy,
A.A.Vartia,
J.W.Burgner,
A.Sutherlin,
V.W.Rodwell,
C.V.Stauffacher.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Biosynthesis of the isoprenoid precursor, isopentenyl diphosphate, is a critical
function in all independently living organisms. There are two major pathways for
this synthesis, the non-mevalonate pathway found in most eubacteria and the
mevalonate pathway found in animal cells and a number of pathogenic bacteria. An
early step in this pathway is the condensation of acetyl-CoA and acetoacetyl-CoA
into HMG-CoA, catalyzed by the enzyme HMG-CoA synthase. To explore the
possibility of a small molecule inhibitor of the enzyme functioning as a
non-cell wall antibiotic, the structure of HMG-CoA synthase from Enterococcus
faecalis (MVAS) was determined by selenomethionine MAD phasing to 2.4 A and the
enzyme complexed with its second substrate, acetoacetyl-CoA, to 1.9 A. These
structures show that HMG-CoA synthase from Enterococcus is a member of the
family of thiolase fold enzymes and, while similar to the recently published
HMG-CoA synthase structures from Staphylococcus aureus, exhibit significant
differences in the structure of the C-terminal domain. The acetoacetyl-CoA
binary structure demonstrates reduced coenzyme A and acetoacetate covalently
bound to the active site cysteine through a thioester bond. This is consistent
with the kinetics of the reaction that have shown acetoacetyl-CoA to be a potent
inhibitor of the overall reaction, and provides a starting point in the search
for a small molecule inhibitor.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
T.J.Buchholz,
C.M.Rath,
N.B.Lopanik,
N.P.Gardner,
K.Håkansson,
and
D.H.Sherman
(2010).
Polyketide β-branching in bryostatin biosynthesis: identification of surrogate acetyl-ACP donors for BryR, an HMG-ACP synthase.
|
| |
Chem Biol,
17,
1092-1100.
|
 |
|
|
|
|
 |
A.M.Haapalainen,
G.Meriläinen,
and
R.K.Wierenga
(2006).
The thiolase superfamily: condensing enzymes with diverse reaction specificities.
|
| |
Trends Biochem Sci,
31,
64-71.
|
 |
|
|
|
|
 |
F.Pojer,
J.L.Ferrer,
S.B.Richard,
D.A.Nagegowda,
M.L.Chye,
T.J.Bach,
and
J.P.Noel
(2006).
Structural basis for the design of potent and species-specific inhibitors of 3-hydroxy-3-methylglutaryl CoA synthases.
|
| |
Proc Natl Acad Sci U S A,
103,
11491-11496.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

