 |
PDBsum entry 1v9e

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.4.2.1.1
- carbonic anhydrase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
hydrogencarbonate + H+ = CO2 + H2O
|
 |
 |
 |
 |
 |
 hydrogencarbonate
hydrogencarbonate
|
+
|
H(+)
|
=
|
 CO2
CO2
|
+
|
H2O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Zn(2+)
|
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.4.2.1.69
- cyanamide hydratase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
urea = cyanamide + H2O
|
 |
 |
 |
 |
 |
 urea
urea
|
=
|
 cyanamide
cyanamide
|
+
|
H2O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Acta Crystallogr D Biol Crystallogr
60:792-795
(2004)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structure of bovine carbonic anhydrase II at 1.95 A resolution.
|
|
R.Saito,
T.Sato,
A.Ikai,
N.Tanaka.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Carbonic anhydrase (CA) is a zinc-containing enzyme that catalyzes the
reversible hydration of CO2 to HCO3-. In eukaryotes, the enzyme plays a role in
various physiological functions, including interconversion between CO2 and HCO3-
in intermediary metabolism, facilitated diffusion of CO2, pH homeostasis and ion
transport. The structure of bovine carbonic anhydrase II (BCA II) has been
determined by molecular replacement and refined to 1.95 A resolution by
simulated-annealing and individual B-factor refinement. The final R factor for
the BCA II structure was 19.4%. BCA II has a C-terminal knot structure similar
to that observed in human CA II. It contains one zinc ion in the active site
coordinated to three histidines and one putative water molecule in a tetrahedral
geometry. The structure of BCA II reveals a probable alternative proton-wire
pathway that differs from that of HCA II.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |
Figure 2.
Figure 2 Schematic drawing of interactions and distances around
the active site determined from the crystal structure of BCA II
at pH 7.5. The net charges assigned by theoretical calculations
using MOPAC are shown. The side-chain atom Gln91 O  1
(net charge -0.30) accepts a hydrogen bond from of His93 N 1
(net charge -0.30) accepts a hydrogen bond from of His93 N 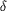 1
(net charge -0.21) and contributes to His93 ligand
stabilization. The side-chain atom of Gln91 N 1
(net charge -0.21) and contributes to His93 ligand
stabilization. The side-chain atom of Gln91 N  2,
with a net charge of -0.40, has a dominant role in the binding
of water molecule W482, with net charge of +0.02, in its
slightly acidic or cationic form. This interaction is likely to
be more hydrogen-bonding in character than the interaction
between W162 (net charge +0.01) and His63 N 2,
with a net charge of -0.40, has a dominant role in the binding
of water molecule W482, with net charge of +0.02, in its
slightly acidic or cationic form. This interaction is likely to
be more hydrogen-bonding in character than the interaction
between W162 (net charge +0.01) and His63 N  2
(net charge -0.14) that has been a biological focus in the case
of HCA II. This finding suggests that the dipole donor group of
Gln91 may also participate in processes that require relatively
rapid proton movement or release. 2
(net charge -0.14) that has been a biological focus in the case
of HCA II. This finding suggests that the dipole donor group of
Gln91 may also participate in processes that require relatively
rapid proton movement or release.
|
 |
|
|

|
| |
The above figure is
reprinted
by permission from the IUCr:
Acta Crystallogr D Biol Crystallogr
(2004,
60,
792-795)
copyright 2004.
|
|
| |
Figure was
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.L.Mallam
(2009).
How does a knotted protein fold?
|
| |
FEBS J,
276,
365-375.
|
 |
|
|
|
|
 |
R.Chiuri,
G.Maiorano,
A.Rizzello,
L.L.del Mercato,
R.Cingolani,
R.Rinaldi,
M.Maffia,
and
P.P.Pompa
(2009).
Exploring local flexibility/rigidity in psychrophilic and mesophilic carbonic anhydrases.
|
| |
Biophys J,
96,
1586-1596.
|
 |
|
|
|
|
 |
A.Ikai
(2008).
Nanobiomechanics of proteins and biomembrane.
|
| |
Philos Trans R Soc Lond B Biol Sci,
363,
2163-2171.
|
 |
|
|
|
|
 |
J.H.Harvey,
and
D.Trauner
(2008).
Regulating enzymatic activity with a photoswitchable affinity label.
|
| |
Chembiochem,
9,
191-193.
|
 |
|
|
|
|
 |
Q.Zhang,
E.Crosland,
and
D.Fabris
(2008).
Nested Arg-specific bifunctional crosslinkers for MS-based structural analysis of proteins and protein assemblies.
|
| |
Anal Chim Acta,
627,
117-128.
|
 |
|
|
|
|
 |
V.M.Krishnamurthy,
G.K.Kaufman,
A.R.Urbach,
I.Gitlin,
K.L.Gudiksen,
D.B.Weibel,
and
G.M.Whitesides
(2008).
Carbonic anhydrase as a model for biophysical and physical-organic studies of proteins and protein-ligand binding.
|
| |
Chem Rev,
108,
946.
|
 |
|
|
|
|
 |
W.Burkitt,
and
G.O'Connor
(2008).
Assessment of the repeatability and reproducibility of hydrogen/deuterium exchange mass spectrometry measurements.
|
| |
Rapid Commun Mass Spectrom,
22,
3893-3901.
|
 |
|
|
|
|
 |
B.Monterroso,
and
A.P.Minton
(2007).
Effect of high concentration of inert cosolutes on the refolding of an enzyme: carbonic anhydrase B in sucrose and ficoll 70.
|
| |
J Biol Chem,
282,
33452-33458.
|
 |
|
|
|
|
 |
S.Safarian,
F.Bagheri,
A.A.Moosavi-Movahedi,
M.Amanlou,
and
N.Sheibani
(2007).
Competitive inhibitory effects of acetazolamide upon interactions with bovine carbonic anhydrase II.
|
| |
Protein J,
26,
371-385.
|
 |
|
|
|
|
 |
S.Ohta,
M.T.Alam,
H.Arakawa,
and
A.Ikai
(2004).
Origin of mechanical strength of bovine carbonic anhydrase studied by molecular dynamics simulation.
|
| |
Biophys J,
87,
4007-4020.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

