 |
PDBsum entry 1v8i

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase
|
 |
|
Title:
|
 |
Crystal structure analysis of the adp-ribose pyrophosphatase
|
|
Structure:
|
 |
Adp-ribose pyrophosphatase. Chain: a. Engineered: yes
|
|
Source:
|
 |
Thermus thermophilus. Organism_taxid: 274. Expressed in: escherichia coli bl21(de3). Expression_system_taxid: 469008.
|
|
Biol. unit:
|
 |
 Dimer (from PDB file)
Dimer (from PDB file)
|
|
Resolution:
|
 |
|
1.76Å
|
R-factor:
|
0.213
|
R-free:
|
0.236
|
|
|
Authors:
|
 |
S.Yoshiba,T.Ooga,N.Nakagawa,T.Shibata,Y.Inoue,S.Yokoyama,S.Kuramitsu, R.Masui,Riken Structural Genomics/proteomics Initiative (Rsgi)
|
Key ref:

|
 |
S.Yoshiba
et al.
(2004).
Structural insights into the Thermus thermophilus ADP-ribose pyrophosphatase mechanism via crystal structures with the bound substrate and metal.
J Biol Chem,
279,
37163-37174.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
09-Jan-04
|
Release date:
|
19-Oct-04
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q84CU3
(Q84CU3_THETH) -
ADP-ribose pyrophosphatase from Thermus thermophilus
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
170 a.a.
150 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.6.1.13
- ADP-ribose diphosphatase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
ADP-D-ribose + H2O = D-ribose 5-phosphate + AMP + 2 H+
|
 |
 |
 |
 |
 |
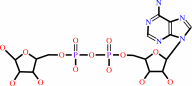 ADP-D-ribose
ADP-D-ribose
|
+
|
H2O
|
=
|
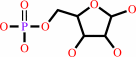 D-ribose 5-phosphate
D-ribose 5-phosphate
|
+
|
 AMP
AMP
|
+
|
2
×
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
279:37163-37174
(2004)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural insights into the Thermus thermophilus ADP-ribose pyrophosphatase mechanism via crystal structures with the bound substrate and metal.
|
|
S.Yoshiba,
T.Ooga,
N.Nakagawa,
T.Shibata,
Y.Inoue,
S.Yokoyama,
S.Kuramitsu,
R.Masui.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
ADP-ribose pyrophosphatase (ADPRase) catalyzes the divalent metal ion-dependent
hydrolysis of ADP-ribose to ribose 5'-phosphate and AMP. This enzyme plays a key
role in regulating the intracellular ADP-ribose levels, and prevents
nonenzymatic ADP-ribosylation. To elucidate the pyrophosphatase hydrolysis
mechanism employed by this enzyme, structural changes occurring on binding of
substrate, metal and product were investigated using crystal structures of
ADPRase from an extreme thermophile, Thermus thermophilus HB8. Seven structures
were determined, including that of the free enzyme, the Zn(2+)-bound enzyme, the
binary complex with ADP-ribose, the ternary complexes with ADP-ribose and Zn(2+)
or Gd(3+), and the product complexes with AMP and Mg(2+) or with ribose
5'-phosphate and Zn(2+). The structural and functional studies suggested that
the ADP-ribose hydrolysis pathway consists of four reaction states: bound with
metal (I), metal and substrate (II), metal and substrate in the transition state
(III), and products (IV). In reaction state II, Glu-82 and Glu-70 abstract a
proton from a water molecule. This water molecule is situated at an ideal
position to carry out nucleophilic attack on the adenosyl phosphate, as it is
3.6 A away from the target phosphorus and almost in line with the scissile bond.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 4.
FIG. 4. Metal binding site. The electron densities of Zn2+
ions (A and B) and Gd^3+ ions (C) derived from the anomalous
difference Fourier maps at peak wavelength are superimposed on
the models. Metal ions are shown as green balls (Zn) or light
green balls (Gd). Water molecules are represented by WA to WD.
The geometries of metal coordination are drawn with blue lines,
and the distances to the metal ion are shown in Table V.
|
 |
Figure 8.
FIG. 8. Schematic representation of the reaction mechanism
of ADPR hydrolysis. The reaction scheme was proposed based on
the active site architecture according to the structural and
functional analyses of TtADPRase. Reaction state I is the
metal-bound state, II is the ternary complex with substrate and
metal, III is the transition state, and IV is the product
release state. The nucleophilic attack and proton abstraction
events are shown by pink arrows.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2004,
279,
37163-37174)
copyright 2004.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
T.Nakamura,
S.Meshitsuka,
S.Kitagawa,
N.Abe,
J.Yamada,
T.Ishino,
H.Nakano,
T.Tsuzuki,
T.Doi,
Y.Kobayashi,
S.Fujii,
M.Sekiguchi,
and
Y.Yamagata
(2010).
Structural and dynamic features of the MutT protein in the recognition of nucleotides with the mutagenic 8-oxoguanine base.
|
| |
J Biol Chem,
285,
444-452.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Coseno,
G.Martin,
C.Berger,
G.Gilmartin,
W.Keller,
and
S.Doublié
(2008).
Crystal structure of the 25 kDa subunit of human cleavage factor Im.
|
| |
Nucleic Acids Res,
36,
3474-3483.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
N.Huang,
L.Sorci,
X.Zhang,
C.A.Brautigam,
X.Li,
N.Raffaelli,
G.Magni,
N.V.Grishin,
A.L.Osterman,
and
H.Zhang
(2008).
Bifunctional NMN adenylyltransferase/ADP-ribose pyrophosphatase: structure and function in bacterial NAD metabolism.
|
| |
Structure,
16,
196-209.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
T.Wakamatsu,
N.Nakagawa,
S.Kuramitsu,
and
R.Masui
(2008).
Structural basis for different substrate specificities of two ADP-ribose pyrophosphatases from Thermus thermophilus HB8.
|
| |
J Bacteriol,
190,
1108-1117.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
G.Ueno,
H.Kanda,
R.Hirose,
K.Ida,
T.Kumasaka,
and
M.Yamamoto
(2006).
RIKEN structural genomics beamlines at the SPring-8; high throughput protein crystallography with automated beamline operation.
|
| |
J Struct Funct Genomics,
7,
15-22.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

