 |
PDBsum entry 1v7l

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.2.1.33
- 3-isopropylmalate dehydratase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Leucine Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(2R,3S)-3-isopropylmalate = (2S)-2-isopropylmalate
|
 |
 |
 |
 |
 |
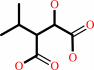 (2R,3S)-3-isopropylmalate
(2R,3S)-3-isopropylmalate
|
=
|
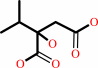 (2S)-2-isopropylmalate
(2S)-2-isopropylmalate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Iron-sulfur
|
 |
 |
 |
 |
 |
 Iron-sulfur
Iron-sulfur
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
344:325-333
(2004)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of the Pyrococcus horikoshii isopropylmalate isomerase small subunit provides insight into the dual substrate specificity of the enzyme.
|
|
Y.Yasutake,
M.Yao,
N.Sakai,
T.Kirita,
I.Tanaka.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Recent studies have implied that the isopropylmalate isomerase small subunit of
the hyperthermophilic archaea Pyrococcus horikoshii (PhIPMI-s) functions as
isopropylmalate isomerase in the leucine biosynthesis pathway, and as
homoaconitase (HACN) in the lysine biosynthesis pathway via alpha-aminoadipic
acid. PhIPMI is thus considered a key to understanding the fundamental
metabolism of the earliest organisms. We describe for the first time the crystal
structure of PhIPMI-s, which displays dual substrate specificity. The crystal
structure unexpectedly shows that four molecules create an interlocked assembly
with intermolecular disulfide linkages having a skewed 222 point-group symmetry.
Although the overall fold of the PhIPMI-s monomer is related closely to domain 4
of the aconitase (ACN), one alpha-helix in the ACN structure is replaced by a
short loop with relatively high temperature factor values. Because this region
is essential for discriminating the structurally similar substrate based on
interactions with its diversified gamma-moiety, the loop structure in the
PhIPMI-s must be dependent on the presence of a substrate. The flexibility of
the loop region might be a structural basis for recognizing both hydrophobic and
hydrophilic gamma-moieties of two distinct substrates, isopropylmalate and
homocitrate.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1. Crystal structure of the PhIPMI-s. (a) Stereoview
ribbon diagram of the PhIPMI-s monomer. The model is colored
according to sequence by a rainbow color ramp from blue at the N
terminus to red at the C terminus. The secondary structures and
the flexible loop moiety are labeled. (b) Superimposition of the
bovine mACN (PDB code, 1ACO) and PhIPMI-s structures. The
PhIPMI-s is colored in green, and the mACN in blue. The
structure of PhIPMI-s is similar to domain 4 of the mACN. The
g-moiety recognition loop of the PhIPMI-s and the corresponding
part of mACN is colored in red. The active-site cleft of mACN is
shown. The superimposition is performed using the program
LSQKAB.33 (c) C^a trace and a diagram of the interlocked
tetramer. The side-chains of the Cys162 forming the
intermolecular disulfide linkages are represented as a
ball-and-stick model. The chains A and B correspond to two
molecules in the asymmetric unit, and chains A' and B' are
generated by the crystallographic 2-fold symmetry. The Figures
were prepared using the programs MOLSCRIPT34 and Raster3D.35
|
 |
Figure 4.
Figure 4. Stereoview of the g-moiety recognition site of
the mACN and PhIPMI-s. (a) Bovine mACN (PDB code, 1ACO). The
structure shows that the Arg580 forms double hydrogen bonds with
the g-carboxylate group of the substrate (aconitate in this
structure). In this diagram, only the residues belonging to
domain 4 are shown. (b) PhIPMI-s. The structure of the mACN in
diagram (a) is shown in gray for comparison. The loop including
Ser65-Arg67 is very well superimposed on the mACN structure,
while the region that would interact with the g-moiety of the
substrate (Tyr24-Lys28) is dissimilar. The helix is replaced by
the loop structure in the PhIPMI-s, and the residues in this
region are absolutely not conserved. The Figures were prepared
using the programs MOLSCRIPT34 and Raster3D.35
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(2004,
344,
325-333)
copyright 2004.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
K.Manikandan,
A.Geerlof,
A.V.Zozulya,
D.I.Svergun,
and
M.S.Weiss
(2011).
Structural studies on the enzyme complex isopropylmalate isomerase (LeuCD) from Mycobacterium tuberculosis.
|
| |
Proteins,
79,
35-49.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
E.M.Larson,
and
A.Idnurm
(2010).
Two origins for the gene encoding alpha-isopropylmalate synthase in fungi.
|
| |
PLoS One,
5,
e11605.
|
 |
|
|
|
|
 |
O.Khersonsky,
and
D.S.Tawfik
(2010).
Enzyme promiscuity: a mechanistic and evolutionary perspective.
|
| |
Annu Rev Biochem,
79,
471-505.
|
 |
|
|
|
|
 |
P.Carbonell,
and
J.L.Faulon
(2010).
Molecular signatures-based prediction of enzyme promiscuity.
|
| |
Bioinformatics,
26,
2012-2019.
|
 |
|
|
|
|
 |
Y.He,
B.Chen,
Q.Pang,
J.M.Strul,
and
S.Chen
(2010).
Functional specification of Arabidopsis isopropylmalate isomerases in glucosinolate and leucine biosynthesis.
|
| |
Plant Cell Physiol,
51,
1480-1487.
|
 |
|
|
|
|
 |
M.Karuppasamy,
A.Geerlof,
L.Schuldt,
C.Mueller-Dieckmann,
and
M.S.Weiss
(2009).
Cloning, expression, purification, crystallization and preliminary X-ray diffraction analysis of the small subunit of isopropylmalate isomerase (Rv2987c) from Mycobacterium tuberculosis.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
65,
136-139.
|
 |
|
|
|
|
 |
T.Knill,
M.Reichelt,
C.Paetz,
J.Gershenzon,
and
S.Binder
(2009).
Arabidopsis thaliana encodes a bacterial-type heterodimeric isopropylmalate isomerase involved in both Leu biosynthesis and the Met chain elongation pathway of glucosinolate formation.
|
| |
Plant Mol Biol,
71,
227-239.
|
 |
|
|
|
|
 |
R.M.Drevland,
A.Waheed,
and
D.E.Graham
(2007).
Enzymology and evolution of the pyruvate pathway to 2-oxobutyrate in Methanocaldococcus jannaschii.
|
| |
J Bacteriol,
189,
4391-4400.
|
 |
|
|
|
|
 |
J.A.McCourt,
and
R.G.Duggleby
(2006).
Acetohydroxyacid synthase and its role in the biosynthetic pathway for branched-chain amino acids.
|
| |
Amino Acids,
31,
173-210.
|
 |
|
|
|
|
 |
J.Dupuy,
A.Volbeda,
P.Carpentier,
C.Darnault,
J.M.Moulis,
and
J.C.Fontecilla-Camps
(2006).
Crystal structure of human iron regulatory protein 1 as cytosolic aconitase.
|
| |
Structure,
14,
129-139.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Yao,
Y.Zhou,
and
I.Tanaka
(2006).
LAFIRE: software for automating the refinement process of protein-structure analysis.
|
| |
Acta Crystallogr D Biol Crystallogr,
62,
189-196.
|
 |
|
|
|
|
 |
P.J.Artymiuk,
and
J.Green
(2006).
The double life of aconitase.
|
| |
Structure,
14,
2-4.
|
 |
|
|
|
|
 |
J.Eichler,
and
M.W.Adams
(2005).
Posttranslational protein modification in Archaea.
|
| |
Microbiol Mol Biol Rev,
69,
393-425.
|
 |
|
|
|
|
 |
K.Hirotsu,
M.Goto,
A.Okamoto,
and
I.Miyahara
(2005).
Dual substrate recognition of aminotransferases.
|
| |
Chem Rec,
5,
160-172.
|
 |
|
|
|
|
 |
N.Leulliot,
S.Quevillon-Cheruel,
M.Graille,
M.Schiltz,
K.Blondeau,
J.Janin,
and
H.Van Tilbeurgh
(2005).
Crystal structure of yeast YER010Cp, a knotable member of the RraA protein family.
|
| |
Protein Sci,
14,
2751-2758.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
Y.Tang,
J.R.Guest,
P.J.Artymiuk,
and
J.Green
(2005).
Switching aconitase B between catalytic and regulatory modes involves iron-dependent dimer formation.
|
| |
Mol Microbiol,
56,
1149-1158.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

