 |
PDBsum entry 1v4n

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
Structure of 5'-deoxy-5'-methylthioadenosine phosphorylase homologue from sulfolobus tokodaii
|
|
Structure:
|
 |
271aa long hypothetical 5'-methylthioadenosine phosphorylase. Chain: a, b, c. Synonym: mtap. Engineered: yes
|
|
Source:
|
 |
Sulfolobus tokodaii. Organism_taxid: 111955. Gene: st0485. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Biol. unit:
|
 |
Trimer (from
)
|
|
Resolution:
|
 |
|
2.45Å
|
R-factor:
|
0.190
|
R-free:
|
0.241
|
|
|
Authors:
|
 |
Y.Kitago,Y.Yasutake,N.Sakai,M.Tsujimura,M.Yao,N.Watanabe, Y.Kawarabayasi,I.Tanaka
|
|
Key ref:
|
 |
Y.Kitago
et al.
Crystal structure of sulfolobus tokodaii mtap.
To be published,
.
|
 |
|
Date:
|
 |
|
14-Nov-03
|
Release date:
|
04-Jan-05
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
F9VN03
(F9VN03_SULTO) -
S-methyl-5'-thioadenosine phosphorylase from Sulfurisphaera tokodaii (strain DSM 16993 / JCM 10545 / NBRC 100140 / 7)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
270 a.a.
266 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.4.2.28
- S-methyl-5'-thioadenosine phosphorylase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
S-methyl-5'-thioadenosine + phosphate = 5-(methylsulfanyl)-alpha-D-ribose 1-phosphate + adenine
|
 |
 |
 |
 |
 |
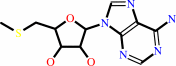 S-methyl-5'-thioadenosine
S-methyl-5'-thioadenosine
|
+
|
 phosphate
phosphate
|
=
|
5-(methylsulfanyl)-alpha-D-ribose 1-phosphate
|
+
|
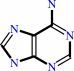 adenine
adenine
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

