 |
PDBsum entry 1tkl

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1tkl
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Yeast oxygen-dependent coproporphyrinogen oxidase
|
|
Structure:
|
 |
Coproporphyrinogen iii oxidase. Chain: a, b. Synonym: coproporphyrinogenase, coprogen oxidase, cox. Engineered: yes
|
|
Source:
|
 |
Saccharomyces cerevisiae. Baker's yeast. Organism_taxid: 4932. Gene: hem13, ydr044w, yd5112.02. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Biol. unit:
|
 |
Dimer (from
)
|
|
Resolution:
|
 |
|
2.00Å
|
R-factor:
|
0.210
|
R-free:
|
0.258
|
|
|
Authors:
|
 |
J.D.Phillips,F.G.Whitby,C.A.Warby,P.Labbe,C.Yang,J.W.Pflugrath, J.D.Ferrara,H.Robinson,J.P.Kushner,C.P.Hill
|
Key ref:

|
 |
J.D.Phillips
et al.
(2004).
Crystal structure of the oxygen-dependant coproporphyrinogen oxidase (Hem13p) of Saccharomyces cerevisiae.
J Biol Chem,
279,
38960-38968.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
08-Jun-04
|
Release date:
|
20-Jul-04
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P11353
(HEM6_YEAST) -
Oxygen-dependent coproporphyrinogen-III oxidase from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
328 a.a.
325 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.3.3.3
- coproporphyrinogen oxidase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Porphyrin Biosynthesis (later stages)
|
 |
 |
 |
 |
 |
Reaction:
|
 |
coproporphyrinogen III + O2 + 2 H+ = protoporphyrinogen IX + 2 CO2 + 2 H2O
|
 |
 |
 |
 |
 |
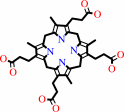 coproporphyrinogen III
coproporphyrinogen III
|
+
|
 O2
O2
|
+
|
2
×
H(+)
|
=
|
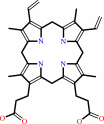 protoporphyrinogen IX
protoporphyrinogen IX
|
+
|
 2
×
CO2
2
×
CO2
|
+
|
2
×
H2O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Fe cation
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
279:38960-38968
(2004)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of the oxygen-dependant coproporphyrinogen oxidase (Hem13p) of Saccharomyces cerevisiae.
|
|
J.D.Phillips,
F.G.Whitby,
C.A.Warby,
P.Labbe,
C.Yang,
J.W.Pflugrath,
J.D.Ferrara,
H.Robinson,
J.P.Kushner,
C.P.Hill.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Coproporphyrinogen oxidase (CPO) is an essential enzyme that catalyzes the sixth
step of the heme biosynthetic pathway. Unusually for heme biosynthetic enzymes,
CPO exists in two evolutionarily and mechanistically distinct families, with
eukaryotes and some prokaryotes employing members of the highly conserved
oxygen-dependent CPO family. Here, we report the crystal structure of the
oxygen-dependent CPO from Saccharomyces cerevisiae (Hem13p), which was
determined by optimized sulfur anomalous scattering and refined to a resolution
of 2.0 A. The protein adopts a novel structure that is quite different from
predicted models and features a central flat seven-stranded anti-parallel sheet
that is flanked by helices. The dimeric assembly, which is seen in different
crystal forms, is formed by packing of helices and a short isolated strand that
forms a beta-ladder with its counterpart in the partner subunit. The deep
active-site cleft is lined by conserved residues and has been captured in open
and closed conformations in two different crystal forms. A substratesized cavity
is completely buried in the closed conformation by the approximately 8-A
movement of a helix that forms a lid over the active site. The structure
therefore suggests residues that likely play critical roles in catalysis and
explains the deleterious effect of many of the mutations associated with the
disease hereditary coproporphyria.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
FIG. 1. Schematic of reaction catalyzed by odCPO/Hem13p.
Propionate side chains on the pyrrole A and B rings are
decarboxylated to form vinyl groups and two molecules of carbon
dioxide. Molecular oxygen is converted to hydrogen peroxide,
presumably via abstraction of a hydrogen atom from each of the
propionate/vinyl C-  atoms. atoms.
|
 |
Figure 6.
FIG. 6. Locations of mutations identified in coproporphyria
patients. Shown is a worm representation stereo view of form II
(closed) structure in the same orientation as shown in Fig. 3A.
Sites of mutations identified in patients are shown as spheres.
Substitutions expected to destabilize the folded protein
structure are shown in gray. Mutations whose presumed
deleterious effect is not easily explained by the structure are
shown in blue. Mutations at the active-site cleft are shown in
magenta. The modeled substrate molecule (white) indicates the
approximate location of the active-site cavity.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2004,
279,
38960-38968)
copyright 2004.
|
|
| |
Figures were
selected
by the author.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
G.Layer,
J.Reichelt,
D.Jahn,
and
D.W.Heinz
(2010).
Structure and function of enzymes in heme biosynthesis.
|
| |
Protein Sci,
19,
1137-1161.
|
 |
|
|
|
|
 |
T.Goto,
R.Aoki,
K.Minamizaki,
and
Y.Fujita
(2010).
Functional differentiation of two analogous coproporphyrinogen III oxidases for heme and chlorophyll biosynthesis pathways in the cyanobacterium Synechocystis sp. PCC 6803.
|
| |
Plant Cell Physiol,
51,
650-663.
|
 |
|
|
|
|
 |
A.Masoumi,
I.U.Heinemann,
M.Rohde,
M.Koch,
M.Jahn,
and
D.Jahn
(2008).
Complex formation between protoporphyrinogen IX oxidase and ferrochelatase during haem biosynthesis in Thermosynechococcus elongatus.
|
| |
Microbiology,
154,
3707-3714.
|
 |
|
|
|
|
 |
P.J.Silva
(2008).
Assessing the reliability of sequence similarities detected through hydrophobic cluster analysis.
|
| |
Proteins,
70,
1588-1594.
|
 |
|
|
|
|
 |
T.Masuda,
and
Y.Fujita
(2008).
Regulation and evolution of chlorophyll metabolism.
|
| |
Photochem Photobiol Sci,
7,
1131-1149.
|
 |
|
|
|
|
 |
J.Fan,
Q.Liu,
Q.Hao,
M.Teng,
and
L.Niu
(2007).
Crystal structure of uroporphyrinogen decarboxylase from Bacillus subtilis.
|
| |
J Bacteriol,
189,
3573-3580.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.R.Stephenson,
J.A.Stacey,
J.B.Morgenthaler,
J.A.Friesen,
T.D.Lash,
and
M.A.Jones
(2007).
Role of aspartate 400, arginine 262, and arginine 401 in the catalytic mechanism of human coproporphyrinogen oxidase.
|
| |
Protein Sci,
16,
401-410.
|
 |
|
|
|
|
 |
A.Andreeva,
and
A.G.Murzin
(2006).
Evolution of protein fold in the presence of functional constraints.
|
| |
Curr Opin Struct Biol,
16,
399-408.
|
 |
|
|
|
|
 |
N.Watanabe
(2006).
From phasing to structure refinement in-house: Cr/Cu dual-wavelength system and a loopless free crystal-mounting method.
|
| |
Acta Crystallogr D Biol Crystallogr,
62,
891-896.
|
 |
|
|
|
|
 |
R.Akagi,
R.Inoue,
S.Muranaka,
T.Tahara,
S.Taketani,
K.E.Anderson,
J.D.Phillips,
and
S.Sassa
(2006).
Dual gene defects involving delta-aminolaevulinate dehydratase and coproporphyrinogen oxidase in a porphyria patient.
|
| |
Br J Haematol,
132,
237-243.
|
 |
|
|
|
|
 |
Y.Hagiwara,
M.Sugishima,
Y.Takahashi,
and
K.Fukuyama
(2006).
Crystal structure of phycocyanobilin:ferredoxin oxidoreductase in complex with biliverdin IXalpha, a key enzyme in the biosynthesis of phycocyanobilin.
|
| |
Proc Natl Acad Sci U S A,
103,
27-32.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
C.Mueller-Dieckmann,
S.Panjikar,
P.A.Tucker,
and
M.S.Weiss
(2005).
On the routine use of soft X-rays in macromolecular crystallography. Part III. The optimal data-collection wavelength.
|
| |
Acta Crystallogr D Biol Crystallogr,
61,
1263-1272.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
D.S.Lee,
E.Flachsová,
M.Bodnárová,
B.Demeler,
P.Martásek,
and
C.S.Raman
(2005).
Structural basis of hereditary coproporphyria.
|
| |
Proc Natl Acad Sci U S A,
102,
14232-14237.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
H.Xu,
C.Yang,
L.Chen,
I.A.Kataeva,
W.Tempel,
D.Lee,
J.E.Habel,
D.Nguyen,
J.W.Pflugrath,
J.D.Ferrara,
W.B.Arendall,
J.S.Richardson,
D.C.Richardson,
Z.J.Liu,
M.G.Newton,
J.P.Rose,
and
B.C.Wang
(2005).
Away from the edge II: in-house Se-SAS phasing with chromium radiation.
|
| |
Acta Crystallogr D Biol Crystallogr,
61,
960-966.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Blanco,
M.Becerra,
M.I.González-Siso,
and
M.E.Cerdán
(2005).
Functional characterization of KlHEM13, a hypoxic gene of Kluyveromyces lactis.
|
| |
Can J Microbiol,
51,
241-249.
|
 |
|
|
|
|
 |
N.Watanabe,
Y.Kitago,
I.Tanaka,
J.Wang,
Y.Gu,
C.Zheng,
and
H.Fan
(2005).
Comparison of phasing methods for sulfur-SAD using in-house chromium radiation: case studies for standard proteins and a 69 kDa protein.
|
| |
Acta Crystallogr D Biol Crystallogr,
61,
1533-1540.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

