 |
PDBsum entry 2d1e

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
2d1e
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.3.7.5
- phycocyanobilin:ferredoxin oxidoreductase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Biliverdin metabolism
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(2R,3Z)-phycocyanobilin + 4 oxidized [2Fe-2S]-[ferredoxin] = biliverdin IXalpha + 4 reduced [2Fe-2S]-[ferredoxin] + 4 H+
|
 |
 |
 |
 |
 |
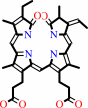 (3Z)-phycocyanobilin
(3Z)-phycocyanobilin
|
+
|
4
×
oxidized ferredoxin
|
=
|
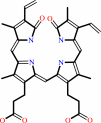 biliverdin IX-alpha
biliverdin IX-alpha
|
+
|
4
×
reduced ferredoxin
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Proc Natl Acad Sci U S A
103:27-32
(2006)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of phycocyanobilin:ferredoxin oxidoreductase in complex with biliverdin IXalpha, a key enzyme in the biosynthesis of phycocyanobilin.
|
|
Y.Hagiwara,
M.Sugishima,
Y.Takahashi,
K.Fukuyama.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Phytobilins (light harvesting and photoreceptor pigments in higher plants,
algae, and cyanobacteria) are synthesized from biliverdin IXalpha (BV) by
ferredoxin-dependent bilin reductases (FDBRs). Phycocyanobilin:ferredoxin
oxidoreductase (PcyA), one such FDBR, is a new class of radical enzymes that
require neither cofactors nor metals and serially reduces the vinyl group of the
D-ring and A-ring of BV using four electrons from ferredoxin to produce
phycocyanobilin, one of the phytobilins. We have determined the crystal
structure of PcyA from Synechocystis sp. PCC 6803 in complex with BV, revealing
the first tertiary structure of an FDBR family member. PcyA is folded in a
three-layer alpha/beta/alpha sandwich structure, in which BV in a cyclic
conformation is positioned between the beta-sheet and C-terminal alpha-helices.
The basic patch on the PcyA surface near the BV molecule may provide a binding
site for acidic ferredoxin, allowing direct transfer of electrons to BV. The
orientation of BV is definitely fixed in PcyA by several hydrophilic
interactions and the shape of the BV binding pocket of PcyA. We propose the
mechanism by which the sequential reduction of the D- and A-rings is controlled,
where Asp-105, located between the two reduction sites, would play the central
role by changing its conformation during the reaction. Homology modeling of
other FDBRs based on the PcyA structure fits well with previous genetic and
biochemical data, thereby providing a structural basis for the reaction
mechanism of FDBRs.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 3.
Fig. 3. Environment of the BV binding site. A schematic
diagram of interactions between PcyA and BV is shown. Dashed
lines indicate hydrogen bonds and salt bridges. The residues
involved in van der Waals interaction with BV are also shown.
For clarity, only the major conformation of Asp-105 is shown.
Carbon, nitrogen, and oxygen atoms are colored in black, blue,
and red, respectively. BV is colored in purple. This figure was
prepared with LIGPLOT (27).
|
 |
Figure 4.
Fig. 4. Proposed reaction mechanism of PcyA. (a) Close-up
view of BV. Omit electron density map for BV (contoured at 2.5
 )
is superimposed on the stick model. The distances between the
carboxyl group of Asp-105 in minor conformation and the lactam
oxygen and nitrogen atoms of D-ring are 3.67 and 2.95 Å,
respectively. (b) Proposed reaction mechanism catalyzed by PcyA.
Reduction schemes at the D-ring (Upper) and at the A-ring
(Lower) are shown. )
is superimposed on the stick model. The distances between the
carboxyl group of Asp-105 in minor conformation and the lactam
oxygen and nitrogen atoms of D-ring are 3.67 and 2.95 Å,
respectively. (b) Proposed reaction mechanism catalyzed by PcyA.
Reduction schemes at the D-ring (Upper) and at the A-ring
(Lower) are shown.
|
 |
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.W.Busch,
E.J.Reijerse,
W.Lubitz,
E.Hofmann,
and
N.Frankenberg-Dinkel
(2011).
Radical mechanism of cyanophage phycoerythrobilin synthase (PebS).
|
| |
Biochem J,
433,
469-476.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.E.Auldridge,
and
K.T.Forest
(2011).
Bacterial phytochromes: More than meets the light.
|
| |
Crit Rev Biochem Mol Biol,
46,
67-88.
|
 |
|
|
|
|
 |
F.Y.Chiu,
Y.R.Chen,
and
S.L.Tu
(2010).
Electrostatic interaction of phytochromobilin synthase and ferredoxin for biosynthesis of phytochrome chromophore.
|
| |
J Biol Chem,
285,
5056-5065.
|
 |
|
|
|
|
 |
Y.Hagiwara,
M.Sugishima,
H.Khawn,
H.Kinoshita,
K.Inomata,
L.Shang,
J.C.Lagarias,
Y.Takahashi,
and
K.Fukuyama
(2010).
Structural insights into vinyl reduction regiospecificity of phycocyanobilin:ferredoxin oxidoreductase (PcyA).
|
| |
J Biol Chem,
285,
1000-1007.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Stoll,
A.Gunn,
M.Brynda,
W.Sughrue,
A.C.Kohler,
A.Ozarowski,
A.J.Fisher,
J.C.Lagarias,
and
R.D.Britt
(2009).
Structure of the biliverdin radical intermediate in phycocyanobilin:ferredoxin oxidoreductase identified by high-field EPR and DFT.
|
| |
J Am Chem Soc,
131,
1986-1995.
|
 |
|
|
|
|
 |
T.Dammeyer,
and
N.Frankenberg-Dinkel
(2008).
Function and distribution of bilin biosynthesis enzymes in photosynthetic organisms.
|
| |
Photochem Photobiol Sci,
7,
1121-1130.
|
 |
|
|
|
|
 |
C.Six,
J.C.Thomas,
L.Garczarek,
M.Ostrowski,
A.Dufresne,
N.Blot,
D.J.Scanlan,
and
F.Partensky
(2007).
Diversity and evolution of phycobilisomes in marine Synechococcus spp. - a comparative genomics study.
|
| |
Genome Biol,
8,
R259.
|
 |
|
|
|
|
 |
N.C.Rockwell,
and
J.C.Lagarias
(2007).
Flexible mapping of homology onto structure with homolmapper.
|
| |
BMC Bioinformatics,
8,
123.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

