 |
PDBsum entry 1td2

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
Crystal structure of the pdxy protein from escherichia coli
|
|
Structure:
|
 |
Pyridoxamine kinase. Chain: a, b. Synonym: pm kinase, pdxy protein. Engineered: yes
|
|
Source:
|
 |
Escherichia coli. Organism_taxid: 562. Gene: pdxy, b1636. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Biol. unit:
|
 |
Dimer (from
)
|
|
Resolution:
|
 |
|
2.22Å
|
R-factor:
|
0.176
|
R-free:
|
0.246
|
|
|
Authors:
|
 |
M.K.Safo,F.N.Musayev,S.Hunt,M.Di Salvo,N.Scarsdale,V.Schirch
|
|
Key ref:
|
 |
M.K.Safo
et al.
(2004).
Crystal structure of the PdxY Protein from Escherichia coli.
J Bacteriol,
186,
8074-8082.
PubMed id:
|
 |
|
Date:
|
 |
|
21-May-04
|
Release date:
|
13-Jul-04
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P77150
(PDXY_ECOLI) -
Pyridoxal kinase PdxY from Escherichia coli (strain K12)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
287 a.a.
287 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.7.1.35
- pyridoxal kinase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
pyridoxal + ATP = pyridoxal 5'-phosphate + ADP + H+
|
 |
 |
 |
 |
 |
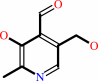 pyridoxal
pyridoxal
|
+
|
ATP
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
 pyridoxal 5'-phosphate
pyridoxal 5'-phosphate
|
+
|
 ADP
ADP
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
J Bacteriol
186:8074-8082
(2004)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of the PdxY Protein from Escherichia coli.
|
|
M.K.Safo,
F.N.Musayev,
S.Hunt,
M.L.di Salvo,
N.Scarsdale,
V.Schirch.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The crystal structure of Escherichia coli PdxY, the protein product of the pdxY
gene, has been determined to a 2.2-A resolution. PdxY is a member of the
ribokinase superfamily of enzymes and has sequence homology with pyridoxal
kinases that phosphorylate pyridoxal at the C-5' hydroxyl. The protein is a
homodimer with an active site on each monomer composed of residues that come
exclusively from each respective subunit. The active site is filled with a
density that fits that of pyridoxal. In monomer A, the ligand appears to be
covalently attached to Cys122 as a thiohemiacetal, while in monomer B it is not
covalently attached but appears to be partially present as pyridoxal
5'-phosphate. The presence of pyridoxal phosphate and pyridoxal as ligands was
confirmed by the activation of aposerine hydroxymethyltransferase after release
of the ligand by the denaturation of PdxY. The ligand, which appears to be
covalently attached to Cys122, does not dissociate after denaturation of the
protein. A detailed comparison (of functional properties, sequence homology,
active site and ATP-binding-site residues, and active site flap types) of PdxY
with other pyridoxal kinases as well as the ribokinase superfamily in general
suggested that PdxY is a member of a new subclass of the ribokinase superfamily.
The structure of PdxY also permitted an interpretation of work that was
previously published about this enzyme.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
C.H.Trinh,
A.Asipu,
D.T.Bonthron,
and
S.E.Phillips
(2009).
Structures of alternatively spliced isoforms of human ketohexokinase.
|
| |
Acta Crystallogr D Biol Crystallogr,
65,
201-211.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
V.Guixé,
and
F.Merino
(2009).
The ADP-dependent sugar kinase family: kinetic and evolutionary aspects.
|
| |
IUBMB Life,
61,
753-761.
|
 |
|
|
|
|
 |
F.Merino,
and
V.Guixé
(2008).
Specificity evolution of the ADP-dependent sugar kinase family: in silico studies of the glucokinase/phosphofructokinase bifunctional enzyme from Methanocaldococcus jannaschii.
|
| |
FEBS J,
275,
4033-4044.
|
 |
|
|
|
|
 |
F.N.Musayev,
M.L.di Salvo,
T.P.Ko,
A.K.Gandhi,
A.Goswami,
V.Schirch,
and
M.K.Safo
(2007).
Crystal Structure of human pyridoxal kinase: structural basis of M(+) and M(2+) activation.
|
| |
Protein Sci,
16,
2184-2194.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.A.Newman,
S.K.Das,
S.E.Sedelnikova,
and
D.W.Rice
(2006).
Cloning, purification and preliminary crystallographic analysis of a putative pyridoxal kinase from Bacillus subtilis.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
62,
1006-1009.
|
 |
|
|
|
|
 |
L.Arnfors,
T.Hansen,
P.Schönheit,
R.Ladenstein,
and
W.Meining
(2006).
Structure of Methanocaldococcus jannaschii nucleoside kinase: an archaeal member of the ribokinase family.
|
| |
Acta Crystallogr D Biol Crystallogr,
62,
1085-1097.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.K.Safo,
F.N.Musayev,
M.L.di Salvo,
S.Hunt,
J.B.Claude,
and
V.Schirch
(2006).
Crystal structure of pyridoxal kinase from the Escherichia coli pdxK gene: implications for the classification of pyridoxal kinases.
|
| |
J Bacteriol,
188,
4542-4552.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

