 |
PDBsum entry 2ddm

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
Crystal structure of pyridoxal kinase from the escherichia coli pdxk gene at 2.1 a resolution
|
|
Structure:
|
 |
Pyridoxine kinase. Chain: a, b. Synonym: pyridoxal kinase, vitamin b6 kinase, pyridoxamine kinase, pn/pl/pm kinase. Engineered: yes
|
|
Source:
|
 |
Escherichia coli. Organism_taxid: 562. Strain: hms 174(de3). Gene: pdxk. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Biol. unit:
|
 |
Dimer (from
)
|
|
Resolution:
|
 |
|
2.10Å
|
R-factor:
|
0.194
|
R-free:
|
0.218
|
|
|
Authors:
|
 |
M.K.Safo,F.N.Musayev,M.L.Di Salvo,S.Hunt,J.B.Claude,V.Schirch
|
|
Key ref:
|
 |
M.K.Safo
et al.
(2006).
Crystal structure of pyridoxal kinase from the Escherichia coli pdxK gene: implications for the classification of pyridoxal kinases.
J Bacteriol,
188,
4542-4552.
PubMed id:
|
 |
|
Date:
|
 |
|
02-Feb-06
|
Release date:
|
15-Aug-06
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P40191
(PDXK_ECOLI) -
Pyridoxine/pyridoxal/pyridoxamine kinase from Escherichia coli (strain K12)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
283 a.a.
264 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.7.1.35
- pyridoxal kinase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
pyridoxal + ATP = pyridoxal 5'-phosphate + ADP + H+
|
 |
 |
 |
 |
 |
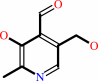 pyridoxal
pyridoxal
|
+
|
ATP
Bound ligand (Het Group name = )
matches with 42.86% similarity
|
=
|
 pyridoxal 5'-phosphate
pyridoxal 5'-phosphate
|
+
|
 ADP
ADP
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
J Bacteriol
188:4542-4552
(2006)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of pyridoxal kinase from the Escherichia coli pdxK gene: implications for the classification of pyridoxal kinases.
|
|
M.K.Safo,
F.N.Musayev,
M.L.di Salvo,
S.Hunt,
J.B.Claude,
V.Schirch.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The pdxK and pdxY genes have been found to code for pyridoxal kinases, enzymes
involved in the pyridoxal phosphate salvage pathway. Two pyridoxal kinase
structures have recently been published, including Escherichia coli pyridoxal
kinase 2 (ePL kinase 2) and sheep pyridoxal kinase, products of the pdxY and
pdxK genes, respectively. We now report the crystal structure of E. coli
pyridoxal kinase 1 (ePL kinase 1), encoded by a pdxK gene, and an isoform of ePL
kinase 2. The structures were determined in the unliganded and binary complexes
with either MgATP or pyridoxal to 2.1-, 2.6-, and 3.2-A resolutions,
respectively. The active site of ePL kinase 1 does not show significant
conformational change upon binding of either pyridoxal or MgATP. Like sheep PL
kinase, ePL kinase 1 exhibits a sequential random mechanism. Unlike sheep
pyridoxal kinase, ePL kinase 1 may not tolerate wide variation in the size and
chemical nature of the 4' substituent on the substrate. This is the result of
differences in a key residue at position 59 on a loop (loop II) that partially
forms the active site. Residue 59, which is His in ePL kinase 1, interacts with
the formyl group at C-4' of pyridoxal and may also determine if residues from
another loop (loop I) can fill the active site in the absence of the substrate.
Both loop I and loop II are suggested to play significant roles in the functions
of PL kinases.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
F.N.Musayev,
M.L.di Salvo,
T.P.Ko,
A.K.Gandhi,
A.Goswami,
V.Schirch,
and
M.K.Safo
(2007).
Crystal Structure of human pyridoxal kinase: structural basis of M(+) and M(2+) activation.
|
| |
Protein Sci,
16,
2184-2194.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.A.Newman,
S.K.Das,
S.E.Sedelnikova,
and
D.W.Rice
(2006).
Cloning, purification and preliminary crystallographic analysis of a putative pyridoxal kinase from Bacillus subtilis.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
62,
1006-1009.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

