 |
PDBsum entry 1t5o

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Translation
|
 |
|
Title:
|
 |
Crystal structure of the translation initiation factor eif-2b, subunit delta, from a. Fulgidus
|
|
Structure:
|
 |
Translation initiation factor eif2b, subunit delta. Chain: a, b, c, d. Synonym: eif2bd
|
|
Source:
|
 |
Archaeoglobus fulgidus. Organism_taxid: 224325. Strain: dsm 4304. Other_details: gene eif2bd
|
|
Biol. unit:
|
 |
 Dimer (from
)
Dimer (from
)
|
|
Resolution:
|
 |
|
1.90Å
|
R-factor:
|
0.197
|
R-free:
|
0.246
|
|
|
Authors:
|
 |
V.N.Malashkevich,S.C.Almo,S.K.Burley,New York Sgx Research Center For Structural Genomics (Nysgxrc)
|
|
Key ref:
|
 |
V.N.Malashkevich
and
s.c.almo
Crystal structure of the translation initiation factor eif-2b, Subunit delta, From a. Fulgidus.
To be published,
.
|
 |
|
Date:
|
 |
|
04-May-04
|
Release date:
|
31-Aug-04
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
O29877
(MTNA_ARCFU) -
Putative methylthioribose-1-phosphate isomerase from Archaeoglobus fulgidus (strain ATCC 49558 / DSM 4304 / JCM 9628 / NBRC 100126 / VC-16)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
339 a.a.
340 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.5.3.1.23
- S-methyl-5-thioribose-1-phosphate isomerase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
5-(methylsulfanyl)-alpha-D-ribose 1-phosphate = 5-(methylsulfanyl)-D- ribulose 1-phosphate
|
 |
 |
 |
 |
 |
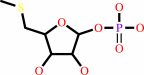 S-methyl-5-thio-alpha-D-ribose 1-phosphate
S-methyl-5-thio-alpha-D-ribose 1-phosphate
|
=
|
5-(methylsulfanyl)ribulose 1-phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

