 |
PDBsum entry 1pym

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Phosphotransferase
|
PDB id
|
|

|

|
1pym
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.5.4.2.9
- phosphoenolpyruvate mutase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Phosphoenolpyruvate Mutase
|
 |
 |
 |
 |
 |
Reaction:
|
 |
phosphoenolpyruvate + H+ = 3-phosphonopyruvate
|
 |
 |
 |
 |
 |
 phosphoenolpyruvate
phosphoenolpyruvate
|
+
|
H(+)
Bound ligand (Het Group name = )
matches with 45.45% similarity
|
=
|
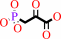 3-phosphonopyruvate
3-phosphonopyruvate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Structure
7:539-548
(1999)
|
|
PubMed id:
|
|
|
|

|
| |
|
Helix swapping between two alpha/beta barrels: crystal structure of phosphoenolpyruvate mutase with bound Mg(2+)-oxalate.
|
|
K.Huang,
Z.Li,
Y.Jia,
D.Dunaway-Mariano,
O.Herzberg.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
BACKGROUND: Phosphonate compounds are important secondary metabolites in nature
and, when linked to macromolecules in eukaryotes, they might play a role in cell
signaling. The first obligatory step in the biosynthesis of phosphonates is the
formation of a carbon-phosphorus bond by converting phosphoenolpyruvate (PEP) to
phosphonopyruvate (P-pyr), a reaction that is catalyzed by PEP mutase. The PEP
mutase functions as a tetramer and requires magnesium ions (Mg2+). RESULTS: The
crystal structure of PEP mutase from the mollusk Mytilus edulis, bound to the
inhibitor Mg(2+)-oxalate, has been determined using multiwavelength anomalous
diffraction, exploiting the selenium absorption edge of a
selenomethionine-containing protein. The structure has been refined at 1.8 A
resolution. PEP mutase adopts a modified alpha/beta barrel fold, in which the
eighth alpha helix projects away from the alpha/beta barrel instead of packing
against the beta sheet. A tightly associated dimer is formed, such that the two
eighth helices are swapped, each packing against the beta sheet of the
neighboring molecule. A dimer of dimers further associates into a tetramer.
Mg(2+)-oxalate is buried close to the center of the barrel, at the C-terminal
ends of the beta strands. CONCLUSIONS: The tetramer observed in the crystal is
likely to be physiologically relevant. Because the Mg(2+)-oxalate is
inaccessible to solvent, substrate binding and dissociation might be accompanied
by conformational changes. A mechanism involving a phosphoenzyme intermediate is
proposed, with Asp58 acting as the nucleophilic entity that accepts and delivers
the phosphoryl group. The active-site architecture and the chemistry performed
by PEP mutase are different from other alpha/beta-barrel proteins that bind
pyruvate or PEP, thus the enzyme might represent a new family of
alpha/beta-barrel proteins.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |
Figure 4.
Figure 4. Cartoon representation of ligand binding to four
α/β-barrel PEP/pyruvate-utilizing enzymes. The β/α motifs
are numbered sequentially. For PEP mutase, helix 8 is colored
yellow because it is contributed by a neighboring molecule. The
loop connecting the β/α motif 3 in pyruvate kinase corresponds
to a whole domain and is shown as a broken yellow line. Motifs 1
and 2 in enolase do not obey the classical topology of β/α
motifs. Residues that interact with PEP/pyruvate/oxalate are
colored red. Residues that interact with Mg^2+ are colored blue.
|
 |
|
|

|
| |
The above figure is
reprinted
by permission from Cell Press:
Structure
(1999,
7,
539-548)
copyright 1999.
|
|
| |
Figure was
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
W.W.Metcalf,
and
W.A.van der Donk
(2009).
Biosynthesis of phosphonic and phosphinic acid natural products.
|
| |
Annu Rev Biochem,
78,
65-94.
|
 |
|
|
|
|
 |
Z.Diaz,
K.B.Xavier,
and
S.T.Miller
(2009).
The crystal structure of the Escherichia coli autoinducer-2 processing protein LsrF.
|
| |
PLoS One,
4,
e6820.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
B.C.Narayanan,
W.Niu,
Y.Han,
J.Zou,
P.S.Mariano,
D.Dunaway-Mariano,
and
O.Herzberg
(2008).
Structure and function of PA4872 from Pseudomonas aeruginosa, a novel class of oxaloacetate decarboxylase from the PEP mutase/isocitrate lyase superfamily.
|
| |
Biochemistry,
47,
167-182.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
C.J.Liao,
K.H.Chin,
C.H.Lin,
P.S.Tsai,
P.C.Lyu,
C.C.Young,
A.H.Wang,
and
S.H.Chou
(2008).
Crystal structure of DFA0005 complexed with alpha-ketoglutarate: a novel member of the ICL/PEPM superfamily from alkali-tolerant Deinococcus ficus.
|
| |
Proteins,
73,
362-371.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
Y.Han,
H.J.Joosten,
W.Niu,
Z.Zhao,
P.S.Mariano,
M.McCalman,
J.van Kan,
P.J.Schaap,
and
D.Dunaway-Mariano
(2007).
Oxaloacetate hydrolase, the C-C bond lyase of oxalate secreting fungi.
|
| |
J Biol Chem,
282,
9581-9590.
|
 |
|
|
|
|
 |
B.Japelj,
J.P.Waltho,
and
R.Jerala
(2004).
Comparison of backbone dynamics of monomeric and domain-swapped stefin A.
|
| |
Proteins,
54,
500-512.
|
 |
|
|
|
|
 |
T.Ose,
K.Watanabe,
M.Yao,
M.Honma,
H.Oikawa,
and
I.Tanaka
(2004).
Structure of macrophomate synthase.
|
| |
Acta Crystallogr D Biol Crystallogr,
60,
1187-1197.
|
 |
|
|
|
|
 |
B.N.Chaudhuri,
M.R.Sawaya,
C.Y.Kim,
G.S.Waldo,
M.S.Park,
T.C.Terwilliger,
and
T.O.Yeates
(2003).
The crystal structure of the first enzyme in the pantothenate biosynthetic pathway, ketopantoate hydroxymethyltransferase, from M tuberculosis.
|
| |
Structure,
11,
753-764.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
F.von Delft,
T.Inoue,
S.A.Saldanha,
H.H.Ottenhof,
F.Schmitzberger,
L.M.Birch,
V.Dhanaraj,
M.Witty,
A.G.Smith,
T.L.Blundell,
and
C.Abell
(2003).
Structure of E. coli ketopantoate hydroxymethyl transferase complexed with ketopantoate and Mg2+, solved by locating 160 selenomethionine sites.
|
| |
Structure,
11,
985-996.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Sarkar,
C.J.Hamilton,
and
A.H.Fairlamb
(2003).
Properties of phosphoenolpyruvate mutase, the first enzyme in the aminoethylphosphonate biosynthetic pathway in Trypanosoma cruzi.
|
| |
J Biol Chem,
278,
22703-22708.
|
 |
|
|
|
|
 |
L.F.Garcia-Alles,
K.Flükiger,
J.Hewel,
R.Gutknecht,
C.Siebold,
S.Schürch,
and
B.Erni
(2002).
Mechanism-based inhibition of enzyme I of the Escherichia coli phosphotransferase system. Cysteine 502 is an essential residue.
|
| |
J Biol Chem,
277,
6934-6942.
|
 |
|
|
|
|
 |
S.Liu,
Z.Lu,
Y.Jia,
D.Dunaway-Mariano,
and
O.Herzberg
(2002).
Dissociative phosphoryl transfer in PEP mutase catalysis: structure of the enzyme/sulfopyruvate complex and kinetic properties of mutants.
|
| |
Biochemistry,
41,
10270-10276.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
Y.Liu,
and
D.Eisenberg
(2002).
3D domain swapping: as domains continue to swap.
|
| |
Protein Sci,
11,
1285-1299.
|
 |
|
|
|
|
 |
K.L.Britton,
I.S.Abeysinghe,
P.J.Baker,
V.Barynin,
P.Diehl,
S.J.Langridge,
B.A.McFadden,
S.E.Sedelnikova,
T.J.Stillman,
K.Weeradechapon,
and
D.W.Rice
(2001).
The structure and domain organization of Escherichia coli isocitrate lyase.
|
| |
Acta Crystallogr D Biol Crystallogr,
57,
1209-1218.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
K.Britton,
S.Langridge,
P.J.Baker,
K.Weeradechapon,
S.E.Sedelnikova,
J.R.De Lucas,
D.W.Rice,
and
G.Turner
(2000).
The crystal structure and active site location of isocitrate lyase from the fungus Aspergillus nidulans.
|
| |
Structure,
8,
349-362.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

