 |
PDBsum entry 1pbb

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1pbb
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Crystal structures of wild-type p-hydroxybenzoate hydroxylase complexed with 4-aminobenzoate, 2,4-dihydroxybenzoate and 2-hydroxy- 4-aminobenzoate and of the try222ala mutant, complexed with 2- hydroxy-4-aminobenzoate. Evidence for a proton channel and a new binding mode of the flavin ring
|
|
Structure:
|
 |
P-hydroxybenzoate hydroxylase. Chain: a. Engineered: yes
|
|
Source:
|
 |
Pseudomonas fluorescens. Organism_taxid: 294
|
|
Biol. unit:
|
 |
 Dimer (from
)
Dimer (from
)
|
|
Resolution:
|
 |
|
|
Authors:
|
 |
H.A.Schreuder,A.Mattevi,W.G.J.Hol
|
Key ref:

|
 |
H.A.Schreuder
et al.
(1994).
Crystal structures of wild-type p-hydroxybenzoate hydroxylase complexed with 4-aminobenzoate,2,4-dihydroxybenzoate, and 2-hydroxy-4-aminobenzoate and of the Tyr222Ala mutant complexed with 2-hydroxy-4-aminobenzoate. Evidence for a proton channel and a new binding mode of the flavin ring.
Biochemistry,
33,
10161-10170.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
06-Jul-94
|
Release date:
|
30-Sep-94
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P00438
(PHHY_PSEFL) -
p-hydroxybenzoate hydroxylase from Pseudomonas fluorescens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
394 a.a.
391 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.14.13.2
- 4-hydroxybenzoate 3-monooxygenase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Benzoate Metabolism
|
 |
 |
 |
 |
 |
Reaction:
|
 |
4-hydroxybenzoate + NADPH + O2 + H+ = 3,4-dihydroxybenzoate + NADP+ + H2O
|
 |
 |
 |
 |
 |
4-hydroxybenzoate
Bound ligand (Het Group name = )
matches with 90.91% similarity
|
+
|
 NADPH
NADPH
|
+
|
 O2
O2
|
+
|
H(+)
|
=
|
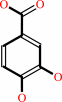 3,4-dihydroxybenzoate
3,4-dihydroxybenzoate
|
+
|
 NADP(+)
NADP(+)
|
+
|
H2O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
FAD
|
 |
 |
 |
 |
 |
FAD
Bound ligand (Het Group name =
FAD)
corresponds exactly
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
33:10161-10170
(1994)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structures of wild-type p-hydroxybenzoate hydroxylase complexed with 4-aminobenzoate,2,4-dihydroxybenzoate, and 2-hydroxy-4-aminobenzoate and of the Tyr222Ala mutant complexed with 2-hydroxy-4-aminobenzoate. Evidence for a proton channel and a new binding mode of the flavin ring.
|
|
H.A.Schreuder,
A.Mattevi,
G.Obmolova,
K.H.Kalk,
W.G.Hol,
F.J.van der Bolt,
W.J.van Berkel.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The crystal structures of wild-type p-hydroxybenzoate hydroxylase from
Pseudomonas fluorescens, complexed with the substrate analogues 4-aminobenzoate,
2,4-dihydroxybenzoate, and 2-hydroxy-4-aminobenzoate have been determined at
2.3-, 2.5-, and 2.8-A resolution, respectively. In addition, the crystal
structure of a Tyr222Ala mutant, complexed with 2-hydroxy-4-aminobenzoate, has
been determined at 2.7-A resolution. The structures have been refined to R
factors between 14.5% and 15.8% for data between 8.0 A and the high-resolution
limit. The differences between these complexes and the wild-type
enzyme-substrate complex are all concentrated in the active site region. Binding
of substrate analogues bearing a 4-amino group (4-aminobenzoate and
2-hydroxy-4-aminobenzoate) leads to binding of a water molecule next to the
active site Tyr385. As a result, a continuous hydrogen-bonding network is
present between the 4-amino group of the substrate analogue and the side chain
of His72. It is likely that this hydrogen-bonding network is transiently present
during normal catalysis, where it may or may not function as a proton channel
assisting the deprotonation of the 4-hydroxyl group of the normal substrate upon
binding to the active site. Binding of substrate analogues bearing a hydroxyl
group at the 2-position (2,4-dihydroxybenzoate and 2-hydroxy-4-aminobenzoate)
leads to displacement of the flavin ring from the active site. The flavin is no
longer in the active site (the "in" conformation) but is in the cleft leading to
the active site instead (the "out" conformation). It is proposed that movement
of the FAD out of the active site may provide an entrance for the substrate to
enter the active site and an exit for the product to leave.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
S.Sah,
and
P.S.Phale
(2011).
1-Naphthol 2-hydroxylase from Pseudomonas sp. strain C6: purification, characterization and chemical modification studies.
|
| |
Biodegradation,
22,
517-526.
|
 |
|
|
|
|
 |
Y.W.Tan,
and
H.Yang
(2011).
Seeing the forest for the trees: fluorescence studies of single enzymes in the context of ensemble experiments.
|
| |
Phys Chem Chem Phys,
13,
1709-1721.
|
 |
|
|
|
|
 |
E.V.Kudryashova,
A.J.Visser,
and
W.J.van Berkel
(2008).
Monomer formation and function of p-hydroxybenzoate hydroxylase in reverse micelles and in dimethylsulfoxide/water mixtures.
|
| |
Chembiochem,
9,
413-419.
|
 |
|
|
|
|
 |
S.Y.Kwon,
B.S.Kang,
G.H.Kim,
and
K.J.Kim
(2007).
Expression, purification, crystallization and initial crystallographic characterization of the p-hydroxybenzoate hydroxylase from Corynebacterium glutamicum.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
63,
944-946.
|
 |
|
|
|
|
 |
T.N.Gustafsson,
T.Sandalova,
J.Lu,
A.Holmgren,
and
G.Schneider
(2007).
High-resolution structures of oxidized and reduced thioredoxin reductase from Helicobacter pylori.
|
| |
Acta Crystallogr D Biol Crystallogr,
63,
833-843.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.H.Westphal,
A.Matorin,
M.A.Hink,
J.W.Borst,
W.J.van Berkel,
and
A.J.Visser
(2006).
Real-time enzyme dynamics illustrated with fluorescence spectroscopy of p-hydroxybenzoate hydroxylase.
|
| |
J Biol Chem,
281,
11074-11081.
|
 |
|
|
|
|
 |
C.Siebold,
N.Berrow,
T.S.Walter,
K.Harlos,
R.J.Owens,
D.I.Stuart,
J.R.Terman,
A.L.Kolodkin,
R.J.Pasterkamp,
and
E.Y.Jones
(2005).
High-resolution structure of the catalytic region of MICAL (molecule interacting with CasL), a multidomain flavoenzyme-signaling molecule.
|
| |
Proc Natl Acad Sci U S A,
102,
16836-16841.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
B.A.Palfey,
Y.V.Murthy,
and
V.Massey
(2003).
Altered balance of half-reactions in p-hydroxybenzoate hydroxylase caused by substituting the 2'-carbon of FAD with fluorine.
|
| |
J Biol Chem,
278,
22210-22216.
|
 |
|
|
|
|
 |
D.Leys,
J.Basran,
and
N.S.Scrutton
(2003).
Channelling and formation of 'active' formaldehyde in dimethylglycine oxidase.
|
| |
EMBO J,
22,
4038-4048.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
U.Kirchner,
A.H.Westphal,
R.Müller,
and
W.J.van Berkel
(2003).
Phenol hydroxylase from Bacillus thermoglucosidasius A7, a two-protein component monooxygenase with a dual role for FAD.
|
| |
J Biol Chem,
278,
47545-47553.
|
 |
|
|
|
|
 |
A.Meyer,
A.Schmid,
M.Held,
A.H.Westphal,
M.Rothlisberger,
H.P.Kohler,
W.J.van Berkel,
and
B.Witholt
(2002).
Changing the substrate reactivity of 2-hydroxybiphenyl 3-monooxygenase from Pseudomonas azelaica HBP1 by directed evolution.
|
| |
J Biol Chem,
277,
5575-5582.
|
 |
|
|
|
|
 |
A.Meyer,
M.Würsten,
A.Schmid,
H.P.Kohler,
and
B.Witholt
(2002).
Hydroxylation of indole by laboratory-evolved 2-hydroxybiphenyl 3-monooxygenase.
|
| |
J Biol Chem,
277,
34161-34167.
|
 |
|
|
|
|
 |
B.A.Palfey,
R.Basu,
K.K.Frederick,
B.Entsch,
and
D.P.Ballou
(2002).
Role of protein flexibility in the catalytic cycle of p-hydroxybenzoate hydroxylase elucidated by the Pro293Ser mutant.
|
| |
Biochemistry,
41,
8438-8446.
|
 |
|
|
|
|
 |
J.Wang,
M.Ortiz-Maldonado,
B.Entsch,
V.Massey,
D.Ballou,
and
D.L.Gatti
(2002).
Protein and ligand dynamics in 4-hydroxybenzoate hydroxylase.
|
| |
Proc Natl Acad Sci U S A,
99,
608-613.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.D.Altose,
Y.Zheng,
J.Dong,
B.A.Palfey,
and
P.R.Carey
(2001).
Comparing protein-ligand interactions in solution and single crystals by Raman spectroscopy.
|
| |
Proc Natl Acad Sci U S A,
98,
3006-3011.
|
 |
|
|
|
|
 |
M.Ortiz-Maldonado,
D.P.Ballou,
and
V.Massey
(2001).
A rate-limiting conformational change of the flavin in p-hydroxybenzoate hydroxylase is necessary for ligand exchange and catalysis: studies with 8-mercapto- and 8-hydroxy-flavins.
|
| |
Biochemistry,
40,
1091-1101.
|
 |
|
|
|
|
 |
A.A.Raibekas,
K.Fukui,
and
V.Massey
(2000).
Design and properties of human D-amino acid oxidase with covalently attached flavin.
|
| |
Proc Natl Acad Sci U S A,
97,
3089-3093.
|
 |
|
|
|
|
 |
M.H.Eppink,
E.Cammaart,
D.Van Wassenaar,
W.J.Middelhoven,
and
W.J.van Berkel
(2000).
Purification and properties of hydroquinone hydroxylase, a FAD-dependent monooxygenase involved in the catabolism of 4-hydroxybenzoate in Candida parapsilosis CBS604.
|
| |
Eur J Biochem,
267,
6832-6840.
|
 |
|
|
|
|
 |
E.Pessione,
S.Divari,
E.Griva,
M.Cavaletto,
G.L.Rossi,
G.Gilardi,
and
C.Giunta
(1999).
Phenol hydroxylase from Acinetobacter radioresistens is a multicomponent enzyme. Purification and characterization of the reductase moiety.
|
| |
Eur J Biochem,
265,
549-555.
|
 |
|
|
|
|
 |
G.R.Moran,
B.Entsch,
B.A.Palfey,
and
D.P.Ballou
(1999).
Mechanistic insights into p-hydroxybenzoate hydroxylase from studies of the mutant Ser212Ala.
|
| |
Biochemistry,
38,
6292-6299.
|
 |
|
|
|
|
 |
M.Ortiz-Maldonado,
D.Gatti,
D.P.Ballou,
and
V.Massey
(1999).
Structure-function correlations of the reaction of reduced nicotinamide analogues with p-hydroxybenzoate hydroxylase substituted with a series of 8-substituted flavins.
|
| |
Biochemistry,
38,
16636-16647.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Ortiz-Maldonado,
D.P.Ballou,
and
V.Massey
(1999).
Use of free energy relationships to probe the individual steps of hydroxylation of p-hydroxybenzoate hydroxylase: studies with a series of 8-substituted flavins.
|
| |
Biochemistry,
38,
8124-8137.
|
 |
|
|
|
|
 |
W.A.Suske,
W.J.van Berkel,
and
H.P.Kohler
(1999).
Catalytic mechanism of 2-hydroxybiphenyl 3-monooxygenase, a flavoprotein from Pseudomonas azelaica HBP1.
|
| |
J Biol Chem,
274,
33355-33365.
|
 |
|
|
|
|
 |
Y.Zheng,
J.Dong,
B.A.Palfey,
and
P.R.Carey
(1999).
Using Raman spectroscopy to monitor the solvent-exposed and "buried" forms of flavin in p-hydroxybenzoate hydroxylase.
|
| |
Biochemistry,
38,
16727-16732.
|
 |
|
|
|
|
 |
A.Mattevi
(1998).
The PHBH fold: not only flavoenzymes.
|
| |
Biophys Chem,
70,
217-222.
|
 |
|
|
|
|
 |
C.Enroth,
H.Neujahr,
G.Schneider,
and
Y.Lindqvist
(1998).
The crystal structure of phenol hydroxylase in complex with FAD and phenol provides evidence for a concerted conformational change in the enzyme and its cofactor during catalysis.
|
| |
Structure,
6,
605-617.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.H.Eppink,
H.A.Schreuder,
and
W.J.van Berkel
(1998).
Interdomain binding of NADPH in p-hydroxybenzoate hydroxylase as suggested by kinetic, crystallographic and modeling studies of histidine 162 and arginine 269 variants.
|
| |
J Biol Chem,
273,
21031-21039.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.Mattevi,
M.A.Vanoni,
and
B.Curti
(1997).
Structure of D-amino acid oxidase: new insights from an old enzyme.
|
| |
Curr Opin Struct Biol,
7,
804-810.
|
 |
|
|
|
|
 |
F.J.van der Bolt,
R.H.van den Heuvel,
J.Vervoort,
and
W.J.van Berkel
(1997).
19F NMR study on the regiospecificity of hydroxylation of tetrafluoro-4-hydroxybenzoate by wild-type and Y385F p-hydroxybenzoate hydroxylase: evidence for a consecutive oxygenolytic dehalogenation mechanism.
|
| |
Biochemistry,
36,
14192-14201.
|
 |
|
|
|
|
 |
G.R.Moran,
B.Entsch,
B.A.Palfey,
and
D.P.Ballou
(1997).
Electrostatic effects on substrate activation in para-hydroxybenzoate hydroxylase: studies of the mutant lysine 297 methionine.
|
| |
Biochemistry,
36,
7548-7556.
|
 |
|
|
|
|
 |
B.Seibold,
M.Matthes,
M.H.Eppink,
F.Lingens,
W.J.Van Berkel,
and
R.Müller
(1996).
4-Hydroxybenzoate hydroxylase from Pseudomonas sp. CBS3. Purification, characterization, gene cloning, sequence analysis and assignment of structural features determining the coenzyme specificity.
|
| |
Eur J Biochem,
239,
469-478.
|
 |
|
|
|
|
 |
D.L.Gatti,
B.Entsch,
D.P.Ballou,
and
M.L.Ludwig
(1996).
pH-dependent structural changes in the active site of p-hydroxybenzoate hydroxylase point to the importance of proton and water movements during catalysis.
|
| |
Biochemistry,
35,
567-578.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
F.J.van der Bolt,
J.Vervoort,
and
W.J.van Berkel
(1996).
Flavin motion in p-hydroxybenzoate hydroxylase. Substrate and effector specificity of the Tyr22-->Ala mutant.
|
| |
Eur J Biochem,
237,
592-600.
|
 |
|
|
|
|
 |
J.F.Gibrat,
T.Madej,
and
S.H.Bryant
(1996).
Surprising similarities in structure comparison.
|
| |
Curr Opin Struct Biol,
6,
377-385.
|
 |
|
|
|
|
 |
M.H.Eppink,
H.A.Schreuder,
and
W.J.Van Berkel
(1995).
Structure and function of mutant Arg44Lys of 4-hydroxybenzoate hydroxylase implications for NADPH binding.
|
| |
Eur J Biochem,
231,
157-165.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
Y.V.Murthy,
and
V.Massey
(1995).
Chemical modification of the N-10 ribityl side chain of flavins. Effects on properties of flavoprotein disulfide oxidoreductases.
|
| |
J Biol Chem,
270,
28586-28594.
|
 |
|
|
|
|
 |
W.J.van Berkel,
M.H.Eppink,
and
H.A.Schreuder
(1994).
Crystal structure of p-hydroxybenzoate hydroxylase reconstituted with the modified FAD present in alcohol oxidase from methylotrophic yeasts: evidence for an arabinoflavin.
|
| |
Protein Sci,
3,
2245-2253.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

