 |
PDBsum entry 1p4c

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1p4c
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.1.1.3.15
- (S)-2-hydroxy-acid oxidase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a (2S)-2-hydroxycarboxylate + O2 = a 2-oxocarboxylate + H2O2
|
 |
 |
 |
 |
 |
(2S)-2-hydroxycarboxylate
|
+
|
 O2
O2
|
=
|
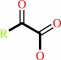 2-oxocarboxylate
2-oxocarboxylate
|
+
|
 H2O2
H2O2
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
FMN
|
 |
 |
 |
 |
 |
FMN
Bound ligand (Het Group name =
FMN)
corresponds exactly
|
|
 |
 |
Enzyme class 2:
|
 |
E.C.1.1.99.31
- (S)-mandelate dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(S)-mandelate + A = phenylglyoxylate + AH2
|
 |
 |
 |
 |
 |
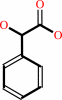 (S)-mandelate
(S)-mandelate
|
+
|
|
=
|
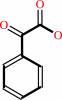 phenylglyoxylate
phenylglyoxylate
|
+
|
AH2
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
279:3749-3757
(2004)
|
|
PubMed id:
|
|
|
|

|
| |
|
High resolution structures of an oxidized and reduced flavoprotein. The water switch in a soluble form of (S)-mandelate dehydrogenase.
|
|
N.Sukumar,
A.R.Dewanti,
B.Mitra,
F.S.Mathews.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The crystal structures of a soluble mutant of the flavoenzyme mandelate
dehydrogenase (MDH) from Pseudomonas putida and of the substrate-reduced enzyme
have been analyzed at 1.35-A resolution. The mutant (MDH-GOX2) is a fully active
chimeric enzyme in which residues 177-215 of the membrane-bound MDH are replaced
by residues 176-195 of glycolate oxidase from spinach. Both structures permit
full tracing of the polypeptide backbone chain from residues 4-356, including a
4-residue segment that was disordered in an earlier study of the oxidized
protein at 2.15 A resolution. The structures of MDH-GOX2 in the oxidized and
reduced states are virtually identical with only a slight increase in the
bending angle of the flavin ring upon reduction. The only other structural
changes within the protein interior are a 10 degrees rotation of an active site
tyrosine side chain, the loss of an active site water, and a significant
movement of six other water molecules in the active site by 0.45 to 0.78 A.
Consistent with solution studies, there is no apparent binding of either the
substrate, mandelate, or the oxidation product, benzoylformate, to the reduced
enzyme. The observed structural changes upon enzyme reduction have been
interpreted as a rearrangement of the hydrogen bonding pattern within the active
site that results from binding of a proton to the N-5 position of the anionic
hydroquinone form of the reduced flavin prosthetic group. Implications for the
low oxidase activity of the reduced enzyme are also discussed.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 4.
FIG. 4. Schematic diagram of the protein environment of FMN
in the oxidized form of MDH. Hydrogen bond distances are in
Å. Residues making hydrophobic contact to FMN are
indicated as shown in the lower right. Carbon atoms are black,
oxygen atoms red, and nitrogen atoms cyan. This diagram was
prepared using the program LIgplot (33)
|
 |
Figure 6.
FIG. 6. Comparison of the oxidized and reduced forms of
MDH-GOX2 at the enzyme active site. The side chains of Tyr26,
Tyr131, backbone of Gly81, the flavin ring, the sulfate anion,
and 7 waters of the oxidized protein are shown in atom colors
(carbon yellow, oxygen red, nitrogen light blue, and sulfur
green). Those atoms of Tyr26 and the 6 waters that show
significant movement, as well as of the sulfate anion in the
reduced enzyme, are shown in dark blue. This diagram was made
using TURBO-FRODO (21).
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2004,
279,
3749-3757)
copyright 2004.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
N.Sukumar,
A.Dewanti,
A.Merli,
G.L.Rossi,
B.Mitra,
and
F.S.Mathews
(2009).
Structures of the G81A mutant form of the active chimera of (S)-mandelate dehydrogenase and its complex with two of its substrates.
|
| |
Acta Crystallogr D Biol Crystallogr,
65,
543-552.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.S.Murray,
R.P.Holmes,
and
W.T.Lowther
(2008).
Active site and loop 4 movements within human glycolate oxidase: implications for substrate specificity and drug design.
|
| |
Biochemistry,
47,
2439-2449.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
I.Leiros,
E.Wang,
T.Rasmussen,
E.Oksanen,
H.Repo,
S.B.Petersen,
P.Heikinheimo,
and
E.Hough
(2006).
The 2.1 A structure of Aerococcus viridans L-lactate oxidase (LOX).
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
62,
1185-1190.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
R.Laupitz,
S.Hecht,
S.Amslinger,
F.Zepeck,
J.Kaiser,
G.Richter,
N.Schramek,
S.Steinbacher,
R.Huber,
D.Arigoni,
A.Bacher,
W.Eisenreich,
and
F.Rohdich
(2004).
Biochemical characterization of Bacillus subtilis type II isopentenyl diphosphate isomerase, and phylogenetic distribution of isoprenoid biosynthesis pathways.
|
| |
Eur J Biochem,
271,
2658-2669.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |
|

