 |
PDBsum entry 1ort

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
Ornithine transcarbamoylase from pseudomonas aeruginosa
|
|
Structure:
|
 |
Ornithine transcarbamoylase. Chain: a, b, c, d, e, f, g, h, i, j, k, l. Synonym: ornithine carbamoyltransferase, otcase. Engineered: yes. Mutation: yes
|
|
Source:
|
 |
Pseudomonas aeruginosa. Organism_taxid: 287
|
|
Biol. unit:
|
 |
Dodecamer (from
)
|
|
Resolution:
|
 |
|
|
Authors:
|
 |
V.Villeret,O.Dideberg
|
|
Key ref:
|
 |
V.Villeret
et al.
(1995).
Crystal structure of Pseudomonas aeruginosa catabolic ornithine transcarbamoylase at 3.0-A resolution: a different oligomeric organization in the transcarbamoylase family.
Proc Natl Acad Sci U S A,
92,
10762-10766.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
24-Aug-95
|
Release date:
|
07-Dec-96
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P08308
(OTCC_PSEAE) -
Ornithine carbamoyltransferase, catabolic from Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
336 a.a.
335 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.1.3.3
- ornithine carbamoyltransferase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Urea Cycle and Arginine Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
carbamoyl phosphate + L-ornithine = L-citrulline + phosphate + H+
|
 |
 |
 |
 |
 |
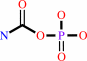 carbamoyl phosphate
carbamoyl phosphate
|
+
|
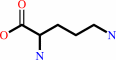 L-ornithine
L-ornithine
|
=
|
 L-citrulline
L-citrulline
|
+
|
 phosphate
phosphate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Proc Natl Acad Sci U S A
92:10762-10766
(1995)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of Pseudomonas aeruginosa catabolic ornithine transcarbamoylase at 3.0-A resolution: a different oligomeric organization in the transcarbamoylase family.
|
|
V.Villeret,
C.Tricot,
V.Stalon,
O.Dideberg.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The crystal structure of the Glu-105-->Gly mutant of catabolic ornithine
transcarbamoylase (OTCase; carbamoyl phosphate + L-ornithine = orthophosphate +
L-citrulline, EC 2.1.3.3) from Pseudomonas aeruginosa has been determined at
3.0-A resolution. This mutant is blocked in the active R (relaxed) state. The
structure was solved by the molecular replacement method, starting from a crude
molecular model built from a trimer of the catalytic subunit of another
transcarbamoylase, the extensively studied aspartate transcarbamoylase (ATCase)
from Escherichia coli. This model was used to generate initial low-resolution
phases at 8-A resolution, which were extended to 3-A by noncrystallographic
symmetry averaging. Four phase extensions were required to obtain an electron
density map of very high quality from which the final model was built. The
structure, including 4020 residues, has been refined to 3-A, and the current
crystallographic R value is 0.216. No solvent molecules have been added to the
model. The catabolic OTCase is a dodecamer composed of four trimers organized in
a tetrahedral manner. Each monomer is composed of two domains. The carbamoyl
phosphate binding domain shows a strong structural homology with the equivalent
ATCase part. In contrast, the other domain, mainly implicated in the binding of
the second substrate (ornithine for OTCase and aspartate for ATCase) is poorly
conserved. The quaternary structures of these two allosteric transcarbamoylases
are quite divergent: the E. coli ATCase has pseudo-32 point-group symmetry, with
six catalytic and six regulatory chains; the catabolic OTCase has 23 point-group
symmetry and only catalytic chains. However, both enzymes display homotropic and
heterotropic cooperativity.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
H.J.Chen,
T.P.Ko,
C.Y.Lee,
N.C.Wang,
and
A.H.Wang
(2009).
Structure, assembly, and mechanism of a PLP-dependent dodecameric L-aspartate beta-decarboxylase.
|
| |
Structure,
17,
517-529.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
B.de Las Rivas,
H.Rodríguez,
I.Angulo,
R.Muñoz,
and
J.M.Mancheño
(2007).
Overexpression, purification, crystallization and preliminary structural studies of catabolic ornithine transcarbamylase from Lactobacillus hilgardii.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
63,
563-567.
|
 |
|
|
|
|
 |
J.Massant,
J.Wouters,
and
N.Glansdorff
(2003).
Refined structure of Pyrococcus furiosus ornithine carbamoyltransferase at 1.87 A.
|
| |
Acta Crystallogr D Biol Crystallogr,
59,
2140-2149.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
Y.Xu,
G.Feller,
C.Gerday,
and
N.Glansdorff
(2003).
Metabolic enzymes from psychrophilic bacteria: challenge of adaptation to low temperatures in ornithine carbamoyltransferase from Moritella abyssi.
|
| |
J Bacteriol,
185,
2161-2168.
|
 |
|
|
|
|
 |
U.Ermler,
C.H.Hagemeier,
A.Roth,
U.Demmer,
W.Grabarse,
E.Warkentin,
and
J.A.Vorholt
(2002).
Structure of methylene-tetrahydromethanopterin dehydrogenase from methylobacterium extorquens AM1.
|
| |
Structure,
10,
1127-1137.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
B.Clantin,
C.Tricot,
T.Lonhienne,
V.Stalon,
and
V.Villeret
(2001).
Probing the role of oligomerization in the high thermal stability of Pyrococcus furiosus ornithine carbamoyltransferase by site-specific mutants.
|
| |
Eur J Biochem,
268,
3937-3942.
|
 |
|
|
|
|
 |
D.Shi,
H.Morizono,
X.Yu,
L.Tong,
N.M.Allewell,
and
M.Tuchman
(2001).
Crystallization and preliminary X-ray crystallographic studies of wild-type human ornithine transcarbamylase and two naturally occurring mutants at position 277.
|
| |
Acta Crystallogr D Biol Crystallogr,
57,
719-721.
|
 |
|
|
|
|
 |
M.Roovers,
R.Sanchez,
C.Legrain,
and
N.Glansdorff
(2001).
Experimental evolution of enzyme temperature activity profile: selection in vivo and characterization of low-temperature-adapted mutants of Pyrococcus furiosus ornithine carbamoyltransferase.
|
| |
J Bacteriol,
183,
1101-1105.
|
 |
|
|
|
|
 |
D.Shi,
H.Morizono,
M.Aoyagi,
M.Tuchman,
and
N.M.Allewell
(2000).
Crystal structure of human ornithine transcarbamylase complexed with carbamoyl phosphate and L-norvaline at 1.9 A resolution.
|
| |
Proteins,
39,
271-277.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
G.Sainz,
J.Vicat,
R.Kahn,
C.Tricot,
V.Stalon,
and
O.Dideberg
(1999).
Purification, crystallization and preliminary X-ray analysis of catabolic ornithine carbamoyltransferase from Pseudomonas aeruginosa.
|
| |
Acta Crystallogr D Biol Crystallogr,
55,
1591-1593.
|
 |
|
|
|
|
 |
F.M.Vellieux
(1998).
A comparison of two algorithms for electron-density map improvement by introduction of atomicity: skeletonization, and map sorting followed by refinement.
|
| |
Acta Crystallogr D Biol Crystallogr,
54,
81-85.
|
 |
|
|
|
|
 |
J.P.Changeux,
and
S.J.Edelstein
(1998).
Allosteric receptors after 30 years.
|
| |
Neuron,
21,
959-980.
|
 |
|
|
|
|
 |
V.Villeret,
B.Clantin,
C.Tricot,
C.Legrain,
M.Roovers,
V.Stalon,
N.Glansdorff,
and
J.Van Beeumen
(1998).
The crystal structure of Pyrococcus furiosus ornithine carbamoyltransferase reveals a key role for oligomerization in enzyme stability at extremely high temperatures.
|
| |
Proc Natl Acad Sci U S A,
95,
2801-2806.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
C.Legrain,
V.Villeret,
M.Roovers,
D.Gigot,
O.Dideberg,
A.Piérard,
and
N.Glansdorff
(1997).
Biochemical characterisation of ornithine carbamoyltransferase from Pyrococcus furiosus.
|
| |
Eur J Biochem,
247,
1046-1055.
|
 |
|
|
|
|
 |
I.D'Hooghe,
C.Vander Wauven,
J.Michiels,
C.Tricot,
P.de Wilde,
J.Vanderleyden,
and
V.Stalon
(1997).
The arginine deiminase pathway in Rhizobium etli: DNA sequence analysis and functional study of the arcABC genes.
|
| |
J Bacteriol,
179,
7403-7409.
|
 |
|
|
|
|
 |
L.Jin,
B.A.Seaton,
and
J.F.Head
(1997).
Crystal structure at 2.8 A resolution of anabolic ornithine transcarbamylase from Escherichia coli.
|
| |
Nat Struct Biol,
4,
622-625.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
R.Sanchez,
M.Baetens,
M.Van de Casteele,
M.Roovers,
C.Legrain,
and
N.Glansdorff
(1997).
Ornithine carbamoyltransferase from the extreme thermophile Thermus thermophilus--analysis of the gene and characterisation of the protein.
|
| |
Eur J Biochem,
248,
466-474.
|
 |
|
|
|
|
 |
Y.Ha,
M.T.McCann,
M.Tuchman,
and
N.M.Allewell
(1997).
Substrate-induced conformational change in a trimeric ornithine transcarbamoylase.
|
| |
Proc Natl Acad Sci U S A,
94,
9550-9555.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
L.B.Murata,
and
H.K.Schachman
(1996).
Structural similarity between ornithine and aspartate transcarbamoylases of Escherichia coli: characterization of the active site and evidence for an interdomain carboxy-terminal helix in ornithine transcarbamoylase.
|
| |
Protein Sci,
5,
709-718.
|
 |
|
|
|
|
 |
N.Mouz,
C.Tricot,
C.Ebel,
Y.Petillot,
V.Stalon,
and
O.Dideberg
(1996).
Use of a designed fusion protein dissociates allosteric properties from the dodecameric state of Pseudomonas aeruginosa catabolic ornithine carbamoyltransferase.
|
| |
Proc Natl Acad Sci U S A,
93,
9414-9419.
|
 |
|
|
|
|
 |
V.T.Nguyen,
D.P.Baker,
C.Tricot,
H.Baur,
V.Villeret,
O.Dideberg,
D.Gigot,
V.Stalon,
and
D.Haas
(1996).
Catabolic ornithine carbamoyltransferase of Pseudomonas aeruginosa. Importance of the N-terminal region for dodecameric structure and homotropic carbamoylphosphate cooperativity.
|
| |
Eur J Biochem,
236,
283-293.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

