 |
PDBsum entry 1mzr

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1mzr
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Structure of dkga from e.Coli at 2.13 a resolution solved by molecular replacement
|
|
Structure:
|
 |
2,5-diketo-d-gluconate reductase a. Chain: a, b. Engineered: yes
|
|
Source:
|
 |
Escherichia coli. Organism_taxid: 562. Gene: yqhe. Expressed in: escherichia coli bl21(de3). Expression_system_taxid: 469008.
|
|
Resolution:
|
 |
|
2.13Å
|
R-factor:
|
0.175
|
R-free:
|
0.220
|
|
|
Authors:
|
 |
C.Abergel,S.Jeudy,V.Monchois,J.M.Claverie,Bacterial Targets At Igs- Cnrs,France (Bigs)
|
Key ref:

|
 |
S.Jeudy
et al.
(2006).
Crystal structure of Escherichia coli DkgA, a broad-specificity aldo-keto reductase.
Proteins,
62,
302-307.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
09-Oct-02
|
Release date:
|
28-Oct-03
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q46857
(DKGA_ECOLI) -
2,5-diketo-D-gluconic acid reductase A from Escherichia coli (strain K12)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
275 a.a.
274 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.1.1.274
- 2,5-didehydrogluconate reductase (2-dehydro-D-gluconate-forming).
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
2-dehydro-D-gluconate + NADP+ = 2,5-didehydro-D-gluconate + NADPH + H+
|
 |
 |
 |
 |
 |
2-dehydro-D-gluconate
Bound ligand (Het Group name = )
matches with 46.15% similarity
|
+
|
 NADP(+)
NADP(+)
|
=
|
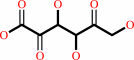 2,5-didehydro-D-gluconate
2,5-didehydro-D-gluconate
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Proteins
62:302-307
(2006)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of Escherichia coli DkgA, a broad-specificity aldo-keto reductase.
|
|
S.Jeudy,
V.Monchois,
C.Maza,
J.M.Claverie,
C.Abergel.
|
|
|

|
| |
ABSTRACT
|
|

|
| |
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1. Stereo view of the ribbon structure of DkgA. It
corresponds to residues 1 to 276. Glycerol and phosphate
molecules are shown in ball-and-stick configuration. The loops
between  4
and 4
and  4, 4,
 7
and H1, and the C-terminal loop surround the entrance of the
active site. 7
and H1, and the C-terminal loop surround the entrance of the
active site.
|
 |
Figure 3.
Figure 3. Activity of DkgA on various substrates. NADPH^+
consumption was followed by the decrease of absorbance at 340
nm. Measurements were performed using 1  M
DkgA and for each substrate a fixed concentration of 5 mM. M
DkgA and for each substrate a fixed concentration of 5 mM.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from John Wiley & Sons, Inc.:
Proteins
(2006,
62,
302-307)
copyright 2006.
|
|
| |
Figures were
selected
by the author.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
E.N.Miller,
L.R.Jarboe,
L.P.Yomano,
S.W.York,
K.T.Shanmugam,
and
L.O.Ingram
(2009).
Silencing of NADPH-dependent oxidoreductase genes (yqhD and dkgA) in furfural-resistant ethanologenic Escherichia coli.
|
| |
Appl Environ Microbiol,
75,
4315-4323.
|
 |
|
|
|
|
 |
J.G.Olsen,
L.Pedersen,
C.L.Christensen,
O.Olsen,
and
A.Henriksen
(2008).
Barley aldose reductase: structure, cofactor binding, and substrate recognition in the aldo/keto reductase 4C family.
|
| |
Proteins,
71,
1572-1581.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

