 |
PDBsum entry 1leh

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1leh
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.4.1.9
- leucine dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-leucine + NAD+ + H2O = 4-methyl-2-oxopentanoate + NH4+ + NADH + H+
|
 |
 |
 |
 |
 |
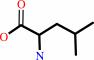 L-leucine
L-leucine
|
+
|
 NAD(+)
NAD(+)
|
+
|
H2O
|
=
|
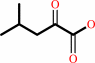 4-methyl-2-oxopentanoate
4-methyl-2-oxopentanoate
|
+
|
NH4(+)
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Structure
3:693-705
(1995)
|
|
PubMed id:
|
|
|
|

|
| |
|
A role for quaternary structure in the substrate specificity of leucine dehydrogenase.
|
|
P.J.Baker,
A.P.Turnbull,
S.E.Sedelnikova,
T.J.Stillman,
D.W.Rice.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
BACKGROUND: Glutamate, phenylalanine and leucine dehydrogenases catalyze the
NAD(P)(+)-linked oxidative deamination of L-amino acids to the corresponding
2-oxoacids, and sequence homology between these enzymes clearly indicates the
existence of an enzyme superfamily related by divergent evolution. We have
undertaken structural studies on a number of members of this family in order to
investigate the molecular basis of their differential amino acid specificity.
RESULTS: We have solved the X-ray structure of the leucine dehydrogenase from
Bacillus sphaericus to a resolution of 2.2 A. Each subunit of this octameric
enzyme contains 364 amino acids and folds into two domains, separated by a deep
cleft. The nicotinamide ring of the NAD+ cofactor binds deep in this cleft,
which is thought to close during the hydride transfer step of the catalytic
cycle. CONCLUSIONS: Comparison of the structure of leucine dehydrogenase with a
hexameric glutamate dehydrogenase has shown that these two enzymes share a
related fold and possess a similar catalytic chemistry. A mechanism for the
basis of the differential amino acid specificity between these enzymes involves
point mutations in the amino acid side-chain specificity pocket and subtle
changes in the shape of this pocket caused by the differences in quaternary
structure.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 2.
Figure 2. Stereo diagrams of a single subunit of LeuDH. The
organization of the subunit into two domains, separated by a
deep cleft, can be seen. In this view the fourfold axis of the
LeuDH octamer runs vertically. (a) Schematic representation with
the strands and helices numbered. (b) Cα trace with every
tenth residue indicated by a black dot. Figure 2. Stereo
diagrams of a single subunit of LeuDH. The organization of the
subunit into two domains, separated by a deep cleft, can be
seen. In this view the fourfold axis of the LeuDH octamer runs
vertically. (a) Schematic representation with the strands and
helices numbered. (b) Cα trace with every tenth residue
indicated by a black dot. (Figure prepared using MOLSCRIPT
[[3]41].)
|
 |
Figure 4.
Figure 4. Stereo ribbon diagram illustrating the interactions
around the fourfold axis in LeuDH. Two monomers are shown (red
and green), viewed down the non-crystallographic twofold axis
which relates pairs of dimers, with the fourfold axis vertical.
Figure 4. Stereo ribbon diagram illustrating the interactions
around the fourfold axis in LeuDH. Two monomers are shown (red
and green), viewed down the non-crystallographic twofold axis
which relates pairs of dimers, with the fourfold axis vertical.
(Figure prepared using FRODO [[3]38].)
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Cell Press:
Structure
(1995,
3,
693-705)
copyright 1995.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
J.Baussand,
and
A.Carbone
(2009).
A combinatorial approach to detect coevolved amino acid networks in protein families of variable divergence.
|
| |
PLoS Comput Biol,
5,
e1000488.
|
 |
|
|
|
|
 |
S.M.Tripathi,
and
R.Ramachandran
(2008).
Crystal structures of the Mycobacterium tuberculosis secretory antigen alanine dehydrogenase (Rv2780) in apo and ternary complex forms captures "open" and "closed" enzyme conformations.
|
| |
Proteins,
72,
1089-1095.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.M.Tripathi,
and
R.Ramachandran
(2008).
Overexpression, purification, crystallization and preliminary X-ray analysis of Rv2780 from Mycobacterium tuberculosis H37Rv.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
64,
367-370.
|
 |
|
|
|
|
 |
S.Y.Seah,
K.L.Britton,
D.W.Rice,
Y.Asano,
and
P.C.Engel
(2003).
Kinetic analysis of phenylalanine dehydrogenase mutants designed for aliphatic amino acid dehydrogenase activity with guidance from homology-based modelling.
|
| |
Eur J Biochem,
270,
4628-4634.
|
 |
|
|
|
|
 |
T.A.Muranova,
S.N.Ruzheinikov,
S.E.Sedelnikova,
P.J.Baker,
A.Pasquo,
A.Galkin,
N.Esaki,
T.Ohshima,
K.Soda,
and
D.W.Rice
(2002).
Crystallization and preliminary X-ray analysis of substrate complexes of leucine dehydrogenase from Thermoactinomyces intermedius.
|
| |
Acta Crystallogr D Biol Crystallogr,
58,
1059-1062.
|
 |
|
|
|
|
 |
T.Oikawa,
K.Yamanaka,
T.Kazuoka,
N.Kanzawa,
and
K.Soda
(2001).
Psychrophilic valine dehydrogenase of the antarctic psychrophile, Cytophaga sp. KUC-1: purification, molecular characterization and expression.
|
| |
Eur J Biochem,
268,
4375-4383.
|
 |
|
|
|
|
 |
X.G.Wang,
K.L.Britton,
T.J.Stillman,
D.W.Rice,
and
P.C.Engel
(2001).
Conversion of a glutamate dehydrogenase into methionine/norleucine dehydrogenase by site-directed mutagenesis.
|
| |
Eur J Biochem,
268,
5791-5799.
|
 |
|
|
|
|
 |
M.Cirilli,
G.Scapin,
A.Sutherland,
J.C.Vederas,
and
J.S.Blanchard
(2000).
The three-dimensional structure of the ternary complex of Corynebacterium glutamicum diaminopimelate dehydrogenase-NADPH-L-2-amino-6-methylene-pimelate.
|
| |
Protein Sci,
9,
2034-2037.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
N.M.Brunhuber,
J.B.Thoden,
J.S.Blanchard,
and
J.L.Vanhooke
(2000).
Rhodococcus L-phenylalanine dehydrogenase: kinetics, mechanism, and structural basis for catalytic specificity.
|
| |
Biochemistry,
39,
9174-9187.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.Sutherland,
and
C.L.Willis
(1999).
Synthesis of probes for the active site of leucine dehydrogenase.
|
| |
Bioorg Med Chem Lett,
9,
1941-1944.
|
 |
|
|
|
|
 |
J.L.Vanhooke,
J.B.Thoden,
N.M.Brunhuber,
J.S.Blanchard,
and
H.M.Holden
(1999).
Phenylalanine dehydrogenase from Rhodococcus sp. M4: high-resolution X-ray analyses of inhibitory ternary complexes reveal key features in the oxidative deamination mechanism.
|
| |
Biochemistry,
38,
2326-2339.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Y.Xu,
G.Bhargava,
H.Wu,
G.Loeber,
and
L.Tong
(1999).
Crystal structure of human mitochondrial NAD(P)+-dependent malic enzyme: a new class of oxidative decarboxylases.
|
| |
Structure,
7,
R877-R889.
|
 |
|
|
|
|
 |
A.Pasquo,
K.L.Britton,
P.J.Baker,
G.Brearley,
R.J.Hinton,
A.J.Moir,
T.J.Stillman,
and
D.W.Rice
(1998).
Crystallization of NAD+-dependent phenylalanine dehydrogenase from Nocardia sp239.
|
| |
Acta Crystallogr D Biol Crystallogr,
54,
269-272.
|
 |
|
|
|
|
 |
P.J.Baker,
Y.Sawa,
H.Shibata,
S.E.Sedelnikova,
and
D.W.Rice
(1998).
Analysis of the structure and substrate binding of Phormidium lapideum alanine dehydrogenase.
|
| |
Nat Struct Biol,
5,
561-567.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Sedelnikova,
D.W.Rice,
H.Shibata,
Y.Sawa,
and
P.J.Baker
(1998).
Crystallization of the alanine dehydrogenase from Phormidium lapideum.
|
| |
Acta Crystallogr D Biol Crystallogr,
54,
407-408.
|
 |
|
|
|
|
 |
A.P.Turnbull,
P.J.Baker,
and
D.W.Rice
(1997).
Analysis of the quaternary structure, substrate specificity, and catalytic mechanism of valine dehydrogenase.
|
| |
J Biol Chem,
272,
25105-25111.
|
 |
|
|
|
|
 |
A.V.Efimov
(1997).
Structural trees for protein superfamilies.
|
| |
Proteins,
28,
241-260.
|
 |
|
|
|
|
 |
G.Scapin,
S.G.Reddy,
and
J.S.Blanchard
(1996).
Three-dimensional structure of meso-diaminopimelic acid dehydrogenase from Corynebacterium glutamicum.
|
| |
Biochemistry,
35,
13540-13551.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

