 |
PDBsum entry 1c1x

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1c1x
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.4.1.20
- phenylalanine dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-phenylalanine + NAD+ + H2O = 3-phenylpyruvate + NH4+ + NADH + H+
|
 |
 |
 |
 |
 |
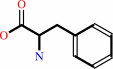 L-phenylalanine
L-phenylalanine
|
+
|
NAD(+)
Bound ligand (Het Group name = )
matches with 84.62% similarity
|
+
|
H2O
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
3-phenylpyruvate
|
+
|
NH4(+)
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Ca(2+)
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
39:9174-9187
(2000)
|
|
PubMed id:
|
|
|
|

|
| |
|
Rhodococcus L-phenylalanine dehydrogenase: kinetics, mechanism, and structural basis for catalytic specificity.
|
|
N.M.Brunhuber,
J.B.Thoden,
J.S.Blanchard,
J.L.Vanhooke.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Phenylalanine dehydrogenase catalyzes the reversible, pyridine
nucleotide-dependent oxidative deamination of L-phenylalanine to form
phenylpyruvate and ammonia. We have characterized the steady-state kinetic
behavior of the enzyme from Rhodococcus sp. M4 and determined the X-ray crystal
structures of the recombinant enzyme in the complexes, E.NADH.L-phenylalanine
and E.NAD(+). L-3-phenyllactate, to 1.25 and 1.4 A resolution, respectively.
Initial velocity, product inhibition, and dead-end inhibition studies indicate
the kinetic mechanism is ordered, with NAD(+) binding prior to phenylalanine and
the products' being released in the order of ammonia, phenylpyruvate, and NADH.
The enzyme shows no activity with NADPH or other 2'-phosphorylated pyridine
nucleotides but has broad activity with NADH analogues. Our initial structural
analyses of the E.NAD(+).phenylpyruvate and E.NAD(+). 3-phenylpropionate
complexes established that Lys78 and Asp118 function as the catalytic residues
in the active site [Vanhooke et al. (1999) Biochemistry 38, 2326-2339]. We have
studied the ionization behavior of these residues in steady-state turnover and
use these findings in conjunction with the structural data described both here
and in our first report to modify our previously proposed mechanism for the
enzymatic reaction. The structural characterizations also illuminate the
mechanism of the redox specificity that precludes alpha-amino acid
dehydrogenases from functioning as alpha-hydroxy acid dehydrogenases.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
C.L.Gerez,
M.S.Carbajo,
G.Rollán,
G.Torres Leal,
and
G.Font de Valdez
(2010).
Inhibition of citrus fungal pathogens by using lactic acid bacteria.
|
| |
J Food Sci,
75,
M354-M359.
|
 |
|
|
|
|
 |
H.F.Fisher,
S.J.Maniscalco,
J.Tally,
and
K.Tabanor
(2009).
Application of the second rule of transient-state kinetic isotope effects to an enzymatic mechanism.
|
| |
Biochemistry,
48,
12265-12271.
|
 |
|
|
|
|
 |
C.A.Bottoms,
P.E.Smith,
and
J.J.Tanner
(2002).
A structurally conserved water molecule in Rossmann dinucleotide-binding domains.
|
| |
Protein Sci,
11,
2125-2137.
|
 |
|
|
|
|
 |
J.F.Tally,
S.J.Maniscalco,
S.K.Saha,
and
H.F.Fisher
(2002).
Detection of multiple active site domain motions in transient-state component time courses of the Clostridium symbiosum L-glutamate dehydrogenase-catalyzed oxidative deamination reaction.
|
| |
Biochemistry,
41,
11284-11293.
|
 |
|
|
|
|
 |
T.A.Muranova,
S.N.Ruzheinikov,
S.E.Sedelnikova,
P.J.Baker,
A.Pasquo,
A.Galkin,
N.Esaki,
T.Ohshima,
K.Soda,
and
D.W.Rice
(2002).
Crystallization and preliminary X-ray analysis of substrate complexes of leucine dehydrogenase from Thermoactinomyces intermedius.
|
| |
Acta Crystallogr D Biol Crystallogr,
58,
1059-1062.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

