 |
PDBsum entry 1kkt

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase
|
 |
|
Title:
|
 |
Structure of p. Citrinum alpha 1,2-mannosidase reveals the basis for differences in specificity of the er and golgi class i enzymes
|
|
Structure:
|
 |
Mannosyl-oligosaccharide alpha-1,2-mannosidase. Chain: a, b. Synonym: man(9)-alpha-mannosidase. Engineered: yes
|
|
Source:
|
 |
Penicillium citrinum. Organism_taxid: 5077. Gene: msdc. Expressed in: aspergillus oryzae. Expression_system_taxid: 5062.
|
|
Resolution:
|
 |
|
2.20Å
|
R-factor:
|
0.193
|
R-free:
|
0.239
|
|
|
Authors:
|
 |
Y.D.Lobsanov,F.Vallee,A.Imberty,T.Yoshida,P.Yip,A.Herscovics, P.L.Howell
|
Key ref:

|
 |
Y.D.Lobsanov
et al.
(2002).
Structure of Penicillium citrinum alpha 1,2-mannosidase reveals the basis for differences in specificity of the endoplasmic reticulum and Golgi class I enzymes.
J Biol Chem,
277,
5620-5630.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
10-Dec-01
|
Release date:
|
23-Jan-02
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P31723
(MAN12_PENCI) -
Mannosyl-oligosaccharide alpha-1,2-mannosidase from Penicillium citrinum
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
511 a.a.
475 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.2.1.113
- mannosyl-oligosaccharide 1,2-alpha-mannosidase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
N4-(alpha-D-Man-(1->2)-alpha-D-Man-(1->2)-alpha-D-Man-(1->3)- [alpha-D-Man-(1->2)-alpha-D-Man-(1->3)-[alpha-D-Man-(1->2)-alpha-D-Man- (1->6)]-alpha-D-Man-(1->6)]-beta-D-Man-(1->4)-beta-D-GlcNAc-(1->4)-beta- D-GlcNAc)-L-asparaginyl-[protein] (N-glucan mannose isomer 9A1,2,3B1,2,3) + 4 H2O = N4-(alpha-D-Man-(1->3)-[alpha-D-Man-(1->3)-[alpha-D-Man- (1->6)]-alpha-D-Man-(1->6)]-beta-D-Man-(1->4)-beta-D-GlcNAc-(1->4)-beta- D-GlcNAc)-L-asparaginyl-[protein] (N-glucan mannose isomer 5A1,2) + 4 beta-D-mannose
|
|
2.
|
N4-(alpha-D-Man-(1->2)-alpha-D-Man-(1->2)-alpha-D-Man-(1->3)- [alpha-D-Man-(1->3)-[alpha-D-Man-(1->2)-alpha-D-Man-(1->6)]-alpha-D-Man- (1->6)]-beta-D-Man-(1->4)-beta-D-GlcNAc-(1->4)-beta-D-GlcNAc)-L- asparaginyl-[protein] (N-glucan mannose isomer 8A1,2,3B1,3) + 3 H2O = N4-(alpha-D-Man-(1->3)-[alpha-D-Man-(1->3)-[alpha-D-Man-(1->6)]-alpha- D-Man-(1->6)]-beta-D-Man-(1->4)-beta-D-GlcNAc-(1->4)-beta-D-GlcNAc)-L- asparaginyl-[protein] (N-glucan mannose isomer 5A1,2) + 3 beta-D-mannose.CC -!- This family of mammalian enzymes, located in the Golgi system,
|
|
 |
 |
 |
 |
 |
N(4)-(alpha-D-Man-(1->2)-alpha-D-Man-(1->2)-alpha-D-Man-(1->3)- [alpha-D-Man-(1->2)-alpha-D-Man-(1->3)-[alpha-D-Man-(1->2)-alpha-D-Man- (1->6)]-alpha-D-Man-(1->6)]-beta-D-Man-(1->4)-beta-D-GlcNAc-(1->4)-beta- D-GlcNAc)-L-asparaginyl-[protein] (N-glucan mannose isomer 9A1,2,3B1,2,3)
|
+
|
4
×
H2O
|
=
|
N(4)-(alpha-D-Man-(1->3)-[alpha-D-Man-(1->3)-[alpha-D-Man- (1->6)]-alpha-D-Man-(1->6)]-beta-D-Man-(1->4)-beta-D-GlcNAc-(1->4)-beta- D-GlcNAc)-L-asparaginyl-[protein] (N-glucan mannose isomer 5A1,2)
Bound ligand (Het Group name = )
matches with 91.67% similarity
|
+
|
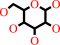 4
×
beta-D-mannose
4
×
beta-D-mannose
|
|
 |
 |
 |
 |
 |
N(4)-(alpha-D-Man-(1->2)-alpha-D-Man-(1->2)-alpha-D-Man-(1->3)- [alpha-D-Man-(1->3)-[alpha-D-Man-(1->2)-alpha-D-Man-(1->6)]-alpha-D-Man- (1->6)]-beta-D-Man-(1->4)-beta-D-GlcNAc-(1->4)-beta-D-GlcNAc)-L- asparaginyl-[protein] (N-glucan mannose isomer 8A1,2,3B1,3)
|
+
|
3
×
H2O
|
=
|
N(4)-(alpha-D-Man-(1->3)-[alpha-D-Man-(1->3)-[alpha-D-Man-(1->6)]-alpha- D-Man-(1->6)]-beta-D-Man-(1->4)-beta-D-GlcNAc-(1->4)-beta-D-GlcNAc)-L- asparaginyl-[protein] (N-glucan mannose isomer 5A1,2)
Bound ligand (Het Group name = )
matches with 91.67% similarity
|
+
|
3
×
beta-D-mannose.CC -!- This family of mammalian enzymes, located in the Golgi system,
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
277:5620-5630
(2002)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structure of Penicillium citrinum alpha 1,2-mannosidase reveals the basis for differences in specificity of the endoplasmic reticulum and Golgi class I enzymes.
|
|
Y.D.Lobsanov,
F.Vallée,
A.Imberty,
T.Yoshida,
P.Yip,
A.Herscovics,
P.L.Howell.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Class I alpha1,2-mannosidases (glycosylhydrolase family 47) are key enzymes in
the maturation of N-glycans. This protein family includes two distinct
enzymatically active subgroups. Subgroup 1 includes the yeast and human
endoplasmic reticulum (ER) alpha1,2-mannosidases that primarily trim
Man(9)GlcNAc(2) to Man(8)GlcNAc(2) isomer B whereas subgroup 2 includes
mammalian Golgi alpha1,2-mannosidases IA, IB, and IC that trim Man(9)GlcNAc(2)
to Man(5)GlcNAc(2) via Man(8)GlcNAc(2) isomers A and C. The structure of the
catalytic domain of the subgroup 2 alpha1,2-mannosidase from Penicillium
citrinum has been determined by molecular replacement at 2.2-A resolution. The
fungal alpha1,2-mannosidase is an (alphaalpha)(7)-helix barrel, very similar to
the subgroup 1 yeast (Vallée, F., Lipari, F., Yip, P., Sleno, B., Herscovics,
A., and Howell, P. L. (2000) EMBO J. 19, 581-588) and human (Vallée, F.,
Karaveg, K., Herscovics, A., Moremen, K. W., and Howell, P. L. (2000) J. Biol.
Chem. 275, 41287-41298) ER enzymes. The location of the conserved acidic
residues of the catalytic site and the binding of the inhibitors, kifunensine
and 1-deoxymannojirimycin, to the essential calcium ion are conserved in the
fungal enzyme. However, there are major structural differences in the
oligosaccharide binding site between the two alpha1,2-mannosidase subgroups. In
the subgroup 1 enzymes, an arginine residue plays a critical role in stabilizing
the oligosaccharide substrate. In the fungal alpha1,2-mannosidase this arginine
is replaced by glycine. This replacement and other sequence variations result in
a more spacious carbohydrate binding site. Modeling studies of interactions
between the yeast, human and fungal enzymes with different Man(8)GlcNAc(2)
isomers indicate that there is a greater degree of freedom to bind the
oligosaccharide in the active site of the fungal enzyme than in the yeast and
human ER alpha1,2-mannosidases.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Fig. 1. Schematic representation of the Man[9]GlcNAc[2].
The mannose residues are labeled M3 to M11. Removal of mannose
residues M11, M10, and M9 results in the formation of
Man[8]GlcNAc[2] isomers A, B, and C, respectively. Residues on
each of the A, B, and C branches are colored purple, red, and
orange, respectively. The torsion angles varied in the energy
calculations, and the corresponding glycosidic linkages are
labeled according to the type of saccharide unit (M, mannose;
GN, N-acetylglucosamine), the chemical linkage (12,  1,2; 13, 1,2; 13,
 1,3; 14, 1,3; 14,
 1,4; 16, 1,4; 16,
 1,6), and
the branch they belong to (A, B, C). To distinguish the 1,6), and
the branch they belong to (A, B, C). To distinguish the  1,2
linkages between M11 and M8 from that between M8 and M5 on
branch A, the linkage between M11 and M8 is designated M12M-AA. 1,2
linkages between M11 and M8 from that between M8 and M5 on
branch A, the linkage between M11 and M8 is designated M12M-AA.
|
 |
Figure 4.
Fig. 4. A, structural superposition of the conserved
acidic residues and calcium ions in the active site region of
the fungal (red), yeast (yellow), and human (blue) enzymes. Only
a subset of the 11 catalytic residues is shown for clarity.
Superposition was done with LSQMAN (34) using all C[  ]atoms.
B, structural superposition of the FM·dMNJ ( pink),
FM·KIF (purple), HM·dMNJ (yellow), and
HM·KIF (green). Only residues implicated in catalysis are
shown. The following fungal residues are shown (the equivalent
yeast numbering is in parentheses): Glu122 (Glu132), Asp267
(Asp275), Ser268 (Ser276), Glu271 (Glu279), Glu409 (Glu435),
Glu412 (Glu438), Glu472 (Glu503), Thr501 (Thr525), Glu502
(Glu526). ]atoms.
B, structural superposition of the FM·dMNJ ( pink),
FM·KIF (purple), HM·dMNJ (yellow), and
HM·KIF (green). Only residues implicated in catalysis are
shown. The following fungal residues are shown (the equivalent
yeast numbering is in parentheses): Glu122 (Glu132), Asp267
(Asp275), Ser268 (Ser276), Glu271 (Glu279), Glu409 (Glu435),
Glu412 (Glu438), Glu472 (Glu503), Thr501 (Thr525), Glu502
(Glu526).
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2002,
277,
5620-5630)
copyright 2002.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
J.Zhou,
C.Z.Lin,
X.Z.Zheng,
X.J.Lin,
W.J.Sang,
S.H.Wang,
Z.H.Wang,
D.Ebbole,
and
G.D.Lu
(2009).
Functional analysis of an alpha-1,2-mannosidase from Magnaporthe oryzae.
|
| |
Curr Genet,
55,
485-496.
|
 |
|
|
|
|
 |
Y.D.Lobsanov,
T.Yoshida,
T.Desmet,
W.Nerinckx,
P.Yip,
M.Claeyssens,
A.Herscovics,
and
P.L.Howell
(2008).
Modulation of activity by Arg407: structure of a fungal alpha-1,2-mannosidase in complex with a substrate analogue.
|
| |
Acta Crystallogr D Biol Crystallogr,
64,
227-236.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.M.Lunetta,
K.A.Simmons,
S.M.Johnson,
and
D.Pappagianis
(2007).
Molecular cloning and expression of a cDNA encoding a Coccidioides posadasii 1,2-alpha-mannosidase identified in the coccidioidal T27K vaccine by immunoproteomic methods.
|
| |
Ann N Y Acad Sci,
1111,
164-180.
|
 |
|
|
|
|
 |
T.Akao,
M.Yamaguchi,
A.Yahara,
K.Yoshiuchi,
H.Fujita,
O.Yamada,
O.Akita,
T.Ohmachi,
Y.Asada,
and
T.Yoshida
(2006).
Cloning and expression of 1,2-alpha-mannosidase gene (fmanIB) from filamentous fungus Aspergillus oryzae: in vivo visualization of the FmanIBp-GFP fusion protein.
|
| |
Biosci Biotechnol Biochem,
70,
471-479.
|
 |
|
|
|
|
 |
Y.Tatara,
B.R.Lee,
T.Yoshida,
K.Takahashi,
and
E.Ichishima
(2003).
Identification of catalytic residues of Ca2+-independent 1,2-alpha-D-mannosidase from Aspergillus saitoi by site-directed mutagenesis.
|
| |
J Biol Chem,
278,
25289-25294.
|
 |
|
|
|
|
 |
A.Varrot,
T.P.Frandsen,
H.Driguez,
and
G.J.Davies
(2002).
Structure of the Humicola insolens cellobiohydrolase Cel6A D416A mutant in complex with a non-hydrolysable substrate analogue, methyl cellobiosyl-4-thio-beta-cellobioside, at 1.9 A.
|
| |
Acta Crystallogr D Biol Crystallogr,
58,
2201-2204.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.Vasella,
G.J.Davies,
and
M.Böhm
(2002).
Glycosidase mechanisms.
|
| |
Curr Opin Chem Biol,
6,
619-629.
|
 |
|
|
|
|
 |
C.Mulakala,
and
P.J.Reilly
(2002).
Understanding protein structure-function relationships in Family 47 alpha-1,2-mannosidases through computational docking of ligands.
|
| |
Proteins,
49,
125-134.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

