 |
PDBsum entry 1kfe

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chains A, B:
E.C.4.2.1.20
- tryptophan synthase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Tryptophan Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(1S,2R)-1-C-(indol-3-yl)glycerol 3-phosphate + L-serine = D-glyceraldehyde 3-phosphate + L-tryptophan + H2O
|
 |
 |
 |
 |
 |
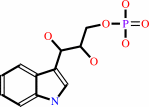 (1S,2R)-1-C-(indol-3-yl)glycerol 3-phosphate
(1S,2R)-1-C-(indol-3-yl)glycerol 3-phosphate
|
+
|
 L-serine
L-serine
|
=
|
 D-glyceraldehyde 3-phosphate
D-glyceraldehyde 3-phosphate
|
+
|
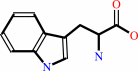 L-tryptophan
L-tryptophan
|
+
|
H2O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
Pyridoxal 5'-phosphate
Bound ligand (Het Group name =
PLS)
matches with 65.22% similarity
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
324:677-690
(2002)
|
|
PubMed id:
|
|
|
|

|
| |
|
On the role of alphaThr183 in the allosteric regulation and catalytic mechanism of tryptophan synthase.
|
|
V.Kulik,
M.Weyand,
R.Seidel,
D.Niks,
D.Arac,
M.F.Dunn,
I.Schlichting.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The catalytic activity and substrate channeling of the pyridoxal
5'-phosphate-dependent tryptophan synthase alpha(2)beta(2) complex is regulated
by allosteric interactions that modulate the switching of the enzyme between
open, low activity and closed, high activity states during the catalytic cycle.
The highly conserved alphaThr183 residue is part of loop alphaL6 and is located
next to the alpha-active site and forms part of the alpha-beta subunit
interface. The role of the interactions of alphaThr183 in alpha-site catalysis
and allosteric regulation was investigated by analyzing the kinetics and crystal
structures of the isosteric mutant alphaThr183Val. The mutant displays strongly
impaired allosteric alpha-beta communication, and the catalytic activity of the
alpha-reaction is reduced one hundred fold, whereas the beta-activity is not
affected. The structural work establishes that the basis for the missing
inter-subunit signaling is the lack of loop alphaL6 closure even in the presence
of the alpha-subunit ligands, 3-indolyl-D-glycerol 3'-phosphate, or
3-indolylpropanol 3'-phosphate. The structural basis for the reduced
alpha-activity has its origins in the missing hydrogen bond between alphaThr183
and the catalytic residue, alphaAsp60.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |
Figure 7.
Figure 7. SigmaA-weighted 2Fo 2 Fc electron density
maps contoured at 1s around the external aldimine
bLys87 in aT183VSer. The Schiff-base linkage between
PLP and bLys87 is broken. As can be seen from Figure
8, bLys87 interacts with the phosphoryl group of PLP.
The Figure was prepared using ``BOBSCRIPT'',
41
MOLS-
CRIPT
39
and RASTER3D.
40
|
 |
|
|

|
| |
The above figure is
reprinted
by permission from Elsevier:
J Mol Biol
(2002,
324,
677-690)
copyright 2002.
|
|
| |
Figure was
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
S.Raboni,
S.Bettati,
and
A.Mozzarelli
(2009).
Tryptophan synthase: a mine for enzymologists.
|
| |
Cell Mol Life Sci,
66,
2391-2403.
|
 |
|
|
|
|
 |
M.F.Dunn,
D.Niks,
H.Ngo,
T.R.Barends,
and
I.Schlichting
(2008).
Tryptophan synthase: the workings of a channeling nanomachine.
|
| |
Trends Biochem Sci,
33,
254-264.
|
 |
|
|
|
|
 |
R.Merkl,
and
M.Zwick
(2008).
H2r: identification of evolutionary important residues by means of an entropy based analysis of multiple sequence alignments.
|
| |
BMC Bioinformatics,
9,
151.
|
 |
|
|
|
|
 |
T.R.Barends,
M.F.Dunn,
and
I.Schlichting
(2008).
Tryptophan synthase, an allosteric molecular factory.
|
| |
Curr Opin Chem Biol,
12,
593-600.
|
 |
|
|
|
|
 |
T.R.Barends,
T.Domratcheva,
V.Kulik,
L.Blumenstein,
D.Niks,
M.F.Dunn,
and
I.Schlichting
(2008).
Structure and mechanistic implications of a tryptophan synthase quinonoid intermediate.
|
| |
Chembiochem,
9,
1024-1028.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.Hardin,
C.F.Villalta,
M.Doan,
M.Jabri,
V.Chockalingham,
S.J.White,
and
R.G.Fowler
(2007).
A molecular characterization of spontaneous frameshift mutagenesis within the trpA gene of Escherichia coli.
|
| |
DNA Repair (Amst),
6,
177-189.
|
 |
|
|
|
|
 |
S.Raboni,
S.Bettati,
and
A.Mozzarelli
(2005).
Identification of the geometric requirements for allosteric communication between the alpha- and beta-subunits of tryptophan synthase.
|
| |
J Biol Chem,
280,
13450-13456.
|
 |
|
|
|
|
 |
F.Schiaretti,
S.Bettati,
C.Viappiani,
and
A.Mozzarelli
(2004).
pH dependence of tryptophan synthase catalytic mechanism: I. The first stage, the beta-elimination reaction.
|
| |
J Biol Chem,
279,
29572-29582.
|
 |
|
|
|
|
 |
Y.Hioki,
K.Ogasahara,
S.J.Lee,
J.Ma,
M.Ishida,
Y.Yamagata,
Y.Matsuura,
M.Ota,
M.Ikeguchi,
S.Kuramitsu,
and
K.Yutani
(2004).
The crystal structure of the tryptophan synthase beta subunit from the hyperthermophile Pyrococcus furiosus. Investigation of stabilization factors.
|
| |
Eur J Biochem,
271,
2624-2635.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |
|

