 |
PDBsum entry 1v8z

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Lyase
|
 |
|
Title:
|
 |
X-ray crystal structure of the tryptophan synthase b2 subunit from hyperthermophile, pyrococcus furiosus
|
|
Structure:
|
 |
Tryptophan synthase beta chain 1. Chain: a, b, c, d. Synonym: tryptophan synthase b2 subunit. Engineered: yes
|
|
Source:
|
 |
Pyrococcus furiosus. Organism_taxid: 2261. Gene: trpb. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Biol. unit:
|
 |
 Dimer (from PDB file)
Dimer (from PDB file)
|
|
Resolution:
|
 |
|
2.21Å
|
R-factor:
|
0.208
|
R-free:
|
0.263
|
|
|
Authors:
|
 |
Y.Hioki,K.Ogasahara,S.J.Lee,J.Ma,M.Ishida,Y.Yamagata,Y.Matsuura, M.Ota,S.Kuramitsu,K.Yutani,Riken Structural Genomics/proteomics Initiative (Rsgi)
|
Key ref:

|
 |
Y.Hioki
et al.
(2004).
The crystal structure of the tryptophan synthase beta subunit from the hyperthermophile Pyrococcus furiosus. Investigation of stabilization factors.
Eur J Biochem,
271,
2624-2635.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
15-Jan-04
|
Release date:
|
22-Feb-05
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q8U093
(TRPB1_PYRFU) -
Tryptophan synthase beta chain 1 from Pyrococcus furiosus (strain ATCC 43587 / DSM 3638 / JCM 8422 / Vc1)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
388 a.a.
386 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.2.1.20
- tryptophan synthase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Tryptophan Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(1S,2R)-1-C-(indol-3-yl)glycerol 3-phosphate + L-serine = D-glyceraldehyde 3-phosphate + L-tryptophan + H2O
|
 |
 |
 |
 |
 |
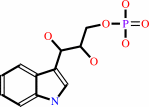 (1S,2R)-1-C-(indol-3-yl)glycerol 3-phosphate
(1S,2R)-1-C-(indol-3-yl)glycerol 3-phosphate
|
+
|
 L-serine
L-serine
|
=
|
 D-glyceraldehyde 3-phosphate
D-glyceraldehyde 3-phosphate
|
+
|
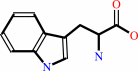 L-tryptophan
L-tryptophan
|
+
|
H2O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
Pyridoxal 5'-phosphate
Bound ligand (Het Group name =
PLP)
matches with 93.75% similarity
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Eur J Biochem
271:2624-2635
(2004)
|
|
PubMed id:
|
|
|
|

|
| |
|
The crystal structure of the tryptophan synthase beta subunit from the hyperthermophile Pyrococcus furiosus. Investigation of stabilization factors.
|
|
Y.Hioki,
K.Ogasahara,
S.J.Lee,
J.Ma,
M.Ishida,
Y.Yamagata,
Y.Matsuura,
M.Ota,
M.Ikeguchi,
S.Kuramitsu,
K.Yutani.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The structure of the tryptophan synthase beta2 subunit (Pfbeta2) from the
hyperthermophile, Pyrococcus furiosus, was determined by X-ray crystallographic
analysis at 2.2 A resolution, and its stability was examined by DSC. This is the
first report of the X-ray structure of the tryptophan synthase beta2 subunit
alone, although the structure of the tryptophan synthase alpha2beta2 complex
from Salmonella typhimurium has already been reported. The structure of Pfbeta2
was essentially similar to that of the beta2 subunit (Stbeta2) in the
alpha2beta2 complex from S. typhimurium. The sequence alignment with secondary
structures of Pfbeta and Stbeta in monomeric form showed that six residues in
the N-terminal region and three residues in the C-terminal region were deleted
in Pfbeta, and one residue at Pro366 of Stbeta and at Ile63 of Pfbeta was
inserted. The denaturation temperature of Pfbeta2 was higher by 35 degrees C
than the reported values from mesophiles at approximately pH 8. On the basis of
structural information on both proteins, the analyses of the contributions of
each stabilization factor indicate that: (a) the higher stability of Pfbeta2 is
not caused by either a hydrophobic interaction or an increase in ion pairs; (b)
the number of hydrogen bonds involved in the main chains of Pfbeta is greater by
about 10% than that of Stbeta, indicating that the secondary structures of
Pfbeta are more stabilized than those of Stbeta and (c) the sequence of Pfbeta
seems to be better fitted to an ideally stable structure than that of Stbeta, as
assessed from X-ray structure data.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 3.
Fig. 3. Crystal structure of  [2] subunit
alone of tryptophan synthase from P. furiosus. (A) The overall
structure of the tryptophan synthase [2] subunit
alone of tryptophan synthase from P. furiosus. (A) The overall
structure of the tryptophan synthase  [2] dimer
from P. furiosus. The N-terminal (1–200) and the C-terminal
(201–388) residues are coloured red and blue, respectively.
Arrows point to the first two strands and one helical structure
(residue 58–64) that intrude into the C domain. The PLP
molecule is represented as a CPK model, coloured gold. Drawings
were prepared using MOLSCRIPT[71]. (B) Two similar N and C
domains of Pf [2] dimer
from P. furiosus. The N-terminal (1–200) and the C-terminal
(201–388) residues are coloured red and blue, respectively.
Arrows point to the first two strands and one helical structure
(residue 58–64) that intrude into the C domain. The PLP
molecule is represented as a CPK model, coloured gold. Drawings
were prepared using MOLSCRIPT[71]. (B) Two similar N and C
domains of Pf  were
superimposed using 69 C were
superimposed using 69 C  pairs fitted
well among the 73 residues of St pairs fitted
well among the 73 residues of St  , which are
reported to deviate by less than 4.0 Å between both
domains [3]. The N and C domains are depicted in gold and green,
respectively. Fitting used program LSQKAB[72]. , which are
reported to deviate by less than 4.0 Å between both
domains [3]. The N and C domains are depicted in gold and green,
respectively. Fitting used program LSQKAB[72].
|
 |
Figure 5.
Fig. 5. Schematic stereo view of the superimposed  monomer
structures of the tryptophan synthase monomer
structures of the tryptophan synthase  [2] from P.
furiosus and S. typhimurium. Blue and red lines represent the
coordinates of Pf [2] from P.
furiosus and S. typhimurium. Blue and red lines represent the
coordinates of Pf  and St and St  (1BKS),
respectively. Drawings were prepared using MOLSCRIPT[71].
Residual numbers are shown with an increase of 10 for the Pf (1BKS),
respectively. Drawings were prepared using MOLSCRIPT[71].
Residual numbers are shown with an increase of 10 for the Pf
 . An arrow
indicates the most different part between the proteins around
position 60 of Pf . An arrow
indicates the most different part between the proteins around
position 60 of Pf  . .
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the Federation of European Biochemical Societies:
Eur J Biochem
(2004,
271,
2624-2635)
copyright 2004.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
K.Nishio,
K.Ogasahara,
Y.Morimoto,
T.Tsukihara,
S.J.Lee,
and
K.Yutani
(2010).
Large conformational changes in the Escherichia coli tryptophan synthase beta(2) subunit upon pyridoxal 5'-phosphate binding.
|
| |
FEBS J,
277,
2157-2170.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Q.Fatmi,
and
C.E.Chang
(2010).
The role of oligomerization and cooperative regulation in protein function: the case of tryptophan synthase.
|
| |
PLoS Comput Biol,
6,
e1000994.
|
 |
|
|
|
|
 |
R.Merkl
(2007).
Modelling the evolution of the archeal tryptophan synthase.
|
| |
BMC Evol Biol,
7,
59.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

