 |
PDBsum entry 1kdg

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1kdg
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.1.99.18
- cellobiose dehydrogenase (acceptor).
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
D-cellobiose + A = D-cellobiono-1,5-lactone + AH2
|
 |
 |
 |
 |
 |
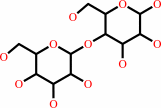 Cellobiose
Cellobiose
|
+
|
 acceptor
acceptor
|
=
|
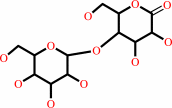 cellobiono-1,5-lactone
cellobiono-1,5-lactone
|
+
|
reduced acceptor
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
FAD; Heme
|
 |
 |
 |
 |
 |
FAD
Bound ligand (Het Group name =
6FA)
matches with 98.15% similarity
|
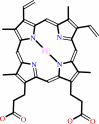 Heme
Heme
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
315:421-434
(2002)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of the flavoprotein domain of the extracellular flavocytochrome cellobiose dehydrogenase.
|
|
B.M.Hallberg,
G.Henriksson,
G.Pettersson,
C.Divne.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Cellobiose dehydrogenase (CDH) participates in the degradation of cellulose and
lignin. The protein is an extracellular flavocytochrome with a b-type cytochrome
domain (CYT(cdh)) connected to a flavodehydrogenase domain (DH(cdh)). DH(cdh)
catalyses a two-electron oxidation at the anomeric C1 position of cellobiose to
yield cellobiono-1,5-lactone, and the electrons are subsequently transferred
from DH(cdh) to an acceptor, either directly or via CYT(cdh). Here, we describe
the crystal structure of Phanerochaete chrysosporium DH(cdh) determined at 1.5 A
resolution. DH(cdh) belongs to the GMC family of oxidoreductases, which includes
glucose oxidase (GOX) and cholesterol oxidase (COX); however, the sequence
identity with members of the family is low. The overall fold of DH(cdh) is
p-hydroxybenzoate hydroxylase-like and is similar to, but also different from,
that of GOX and COX. It is partitioned into an FAD-binding subdomain of
alpha/beta type and a substrate-binding subdomain consisting of a seven-stranded
beta sheet and six helices. Docking of CYT(cdh) and DH(cdh) suggests that
CYT(cdh) covers the active-site entrance in DH(cdh), and that the resulting
distance between the cofactors is within acceptable limits for inter-domain
electron transfer. Based on docking of the substrate, cellobiose, in the active
site of DH(cdh), we propose that the enzyme discriminates against glucose by
favouring interaction with the non-reducing end of cellobiose.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 6.
Figure 6. Comparison of the loop-and-lid structure in COX,
GOX and DH[cdh]. (a) COX[Bre] (loop 46-94; lid 95-109), (b)
GOX[Asp] (loop 54-75; lid 76-97), and (c) DH[cdh] (loop 250-288;
lid residues 289-299). The loop and lid segments are coloured
red and blue, respectively. The flavin cofactor is coloured
yellow. The picture was made as described for Figure 1(a).
|
 |
Figure 7.
Figure 7. Stereo view of the superimposed active sites in
DH[cdh], GOX and COX. Atom colours: carbon (DH[cdh], yellow;
GOX, pink; COX, green); oxygen, red; nitrogen, blue. Residues
Asn309, His689 and Asn732, as well as the isoalloxazine ring of
the FAD cofactor are shown for DH[cdh]. Superposition was made
to obtain optimal alignment of the pyrimidine moiety and the N5
atom of the flavin ring system. The picture was made as
described for Figure 1(a).
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(2002,
315,
421-434)
copyright 2002.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
L.G.Vasilchenko,
K.N.Karapetyan,
O.P.Yershevich,
R.Ludwig,
M.Zamocky,
C.K.Peterbauer,
D.Haltrich,
and
M.L.Rabinovich
(2011).
Cellobiose dehydrogenase of Chaetomium sp. INBI 2-26(-): Structural basis of enhanced activity toward glucose at neutral pH.
|
| |
Biotechnol J,
6,
538-553.
|
 |
|
|
|
|
 |
R.Zhang,
Z.Fan,
and
T.Kasuga
(2011).
Expression of cellobiose dehydrogenase from Neurospora crassa in Pichia pastoris and its purification and characterization.
|
| |
Protein Expr Purif,
75,
63-69.
|
 |
|
|
|
|
 |
Desriani,
S.Ferri,
and
K.Sode
(2010).
Functional expression of Phanerochaete chrysosporium cellobiose dehydrogenase flavin domain in Escherichia coli.
|
| |
Biotechnol Lett,
32,
855-859.
|
 |
|
|
|
|
 |
O.Spadiut,
T.C.Tan,
I.Pisanelli,
D.Haltrich,
and
C.Divne
(2010).
Importance of the gating segment in the substrate-recognition loop of pyranose 2-oxidase.
|
| |
FEBS J,
277,
2892-2909.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
R.Ludwig,
W.Harreither,
F.Tasca,
and
L.Gorton
(2010).
Cellobiose dehydrogenase: a versatile catalyst for electrochemical applications.
|
| |
Chemphyschem,
11,
2674-2697.
|
 |
|
|
|
|
 |
I.S.Fernández,
F.J.Ruíz-Dueñas,
E.Santillana,
P.Ferreira,
M.J.Martínez,
A.T.Martínez,
and
A.Romero
(2009).
Novel structural features in the GMC family of oxidoreductases revealed by the crystal structure of fungal aryl-alcohol oxidase.
|
| |
Acta Crystallogr D Biol Crystallogr,
65,
1196-1205.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
P.Ferreira,
A.Hernandez-Ortega,
B.Herguedas,
A.T.Martínez,
and
M.Medina
(2009).
Aryl-alcohol oxidase involved in lignin degradation: a mechanistic study based on steady and pre-steady state kinetics and primary and solvent isotope effects with two alcohol substrates.
|
| |
J Biol Chem,
284,
24840-24847.
|
 |
|
|
|
|
 |
W.Harreither,
C.Sygmund,
E.Dünhofen,
R.Vicuña,
D.Haltrich,
and
R.Ludwig
(2009).
Cellobiose dehydrogenase from the ligninolytic basidiomycete Ceriporiopsis subvermispora.
|
| |
Appl Environ Microbiol,
75,
2750-2757.
|
 |
|
|
|
|
 |
C.Michalski,
H.Mohagheghi,
M.Nimtz,
J.Pasteels,
and
D.Ober
(2008).
Salicyl alcohol oxidase of the chemical defense secretion of two chrysomelid leaf beetles. Molecular and functional characterization of two new members of the glucose-methanol-choline oxidoreductase gene family.
|
| |
J Biol Chem,
283,
19219-19228.
|
 |
|
|
|
|
 |
P.Baldrian,
and
V.Valásková
(2008).
Degradation of cellulose by basidiomycetous fungi.
|
| |
FEMS Microbiol Rev,
32,
501-521.
|
 |
|
|
|
|
 |
M.Kujawa,
J.Volc,
P.Halada,
P.Sedmera,
C.Divne,
C.Sygmund,
C.Leitner,
C.Peterbauer,
and
D.Haltrich
(2007).
Properties of pyranose dehydrogenase purified from the litter-degrading fungus Agaricus xanthoderma.
|
| |
FEBS J,
274,
879-894.
|
 |
|
|
|
|
 |
M.Kujawa,
H.Ebner,
C.Leitner,
B.M.Hallberg,
M.Prongjit,
J.Sucharitakul,
R.Ludwig,
U.Rudsander,
C.Peterbauer,
P.Chaiyen,
D.Haltrich,
and
C.Divne
(2006).
Structural basis for substrate binding and regioselective oxidation of monosaccharides at C3 by pyranose 2-oxidase.
|
| |
J Biol Chem,
281,
35104-35115.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
P.Ferreira,
F.J.Ruiz-Dueñas,
M.J.Martínez,
W.J.van Berkel,
and
A.T.Martínez
(2006).
Site-directed mutagenesis of selected residues at the active site of aryl-alcohol oxidase, an H2O2-producing ligninolytic enzyme.
|
| |
FEBS J,
273,
4878-4888.
|
 |
|
|
|
|
 |
K.R.Marshall,
M.Gong,
L.Wodke,
J.H.Lamb,
D.J.Jones,
P.B.Farmer,
N.S.Scrutton,
and
A.W.Munro
(2005).
The human apoptosis-inducing protein AMID is an oxidoreductase with a modified flavin cofactor and DNA binding activity.
|
| |
J Biol Chem,
280,
30735-30740.
|
 |
|
|
|
|
 |
L.Stoica,
N.Dimcheva,
D.Haltrich,
T.Ruzgas,
and
L.Gorton
(2005).
Electrochemical investigation of cellobiose dehydrogenase from new fungal sources on Au electrodes.
|
| |
Biosens Bioelectron,
20,
2010-2018.
|
 |
|
|
|
|
 |
T.Kajisa,
M.Yoshida,
K.Igarashi,
A.Katayama,
T.Nishino,
and
M.Samejima
(2004).
Characterization and molecular cloning of cellobiose dehydrogenase from the brown-rot fungus Coniophora puteana.
|
| |
J Biosci Bioeng,
98,
57-63.
|
 |
|
|
|
|
 |
B.M.Hallberg,
G.Henriksson,
G.Pettersson,
A.Vasella,
and
C.Divne
(2003).
Mechanism of the reductive half-reaction in cellobiose dehydrogenase.
|
| |
J Biol Chem,
278,
7160-7166.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Ghanem,
F.Fan,
K.Francis,
and
G.Gadda
(2003).
Spectroscopic and kinetic properties of recombinant choline oxidase from Arthrobacter globiformis.
|
| |
Biochemistry,
42,
15179-15188.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

