 |
PDBsum entry 1jqb

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1jqb
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Alcohol dehydrogenase from clostridium beijerinckii: crystal structure of mutant with enhanced thermal stability
|
|
Structure:
|
 |
NADP-dependent alcohol dehydrogenase. Chain: a, b, c, d. Engineered: yes. Mutation: yes
|
|
Source:
|
 |
Clostridium beijerinckii. Organism_taxid: 1520. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Biol. unit:
|
 |
Tetramer (from
)
|
|
Resolution:
|
 |
|
1.97Å
|
R-factor:
|
0.219
|
R-free:
|
0.247
|
|
|
Authors:
|
 |
I.Levin,F.Frolow,O.Bogin,M.Peretz,Y.Hacham,Y.Burstein
|
Key ref:

|
 |
O.Bogin
et al.
(2002).
Structural basis for the enhanced thermal stability of alcohol dehydrogenase mutants from the mesophilic bacterium Clostridium beijerinckii: contribution of salt bridging.
Protein Sci,
11,
2561-2574.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
05-Aug-01
|
Release date:
|
13-Nov-02
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P25984
(ADH_CLOBE) -
NADP-dependent isopropanol dehydrogenase from Clostridium beijerinckii
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
351 a.a.
351 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 4 residue positions (black
crosses)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.1.1.80
- isopropanol dehydrogenase (NADP(+)).
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
propan-2-ol + NADP+ = acetone + NADPH + H+
|
 |
 |
 |
 |
 |
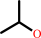 propan-2-ol
propan-2-ol
|
+
|
 NADP(+)
NADP(+)
|
=
|
 acetone
acetone
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Protein Sci
11:2561-2574
(2002)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural basis for the enhanced thermal stability of alcohol dehydrogenase mutants from the mesophilic bacterium Clostridium beijerinckii: contribution of salt bridging.
|
|
O.Bogin,
I.Levin,
Y.Hacham,
S.Tel-Or,
M.Peretz,
F.Frolow,
Y.Burstein.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Previous research in our laboratory comparing the three-dimensional structural
elements of two highly homologous alcohol dehydrogenases, one from the mesophile
Clostridium beijerinckii (CbADH) and the other from the extreme thermophile
Thermoanaerobacter brockii (TbADH), suggested that in the thermophilic enzyme,
an extra intrasubunit ion pair (Glu224-Lys254) and a short ion-pair network
(Lys257-Asp237-Arg304-Glu165) at the intersubunit interface might contribute to
the extreme thermal stability of TbADH. In the present study, we used
site-directed mutagenesis to replace these structurally strategic residues in
CbADH with the corresponding amino acids from TbADH, and we determined the
effect of such replacements on the thermal stability of CbADH. Mutations in the
intrasubunit ion pair region increased thermostability in the single mutant
S254K- and in the double mutant V224E/S254K-CbADH, but not in the single mutant
V224E-CbADH. Both single amino acid replacements, M304R- and Q165E-CbADH, in the
region of the intersubunit ion pair network augmented thermal stability, with an
additive effect in the double mutant M304R/Q165E-CbADH. To investigate the
precise mechanism by which such mutations alter the molecular structure of CbADH
to achieve enhanced thermostability, we constructed a quadruple mutant
V224E/S254K/Q165E/M304R-CbADH and solved its three-dimensional structure. The
overall results indicate that the amino acid substitutions in CbADH mutants with
enhanced thermal stability reinforce the quaternary structure of the enzyme by
formation of an extended network of intersubunit ion pairs and salt bridges,
mediated by water molecules, and by forming a new intrasubunit salt bridge.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 3.
Fig. 3. Three-dimensional structure of the tetramer of the
quadruple mutant of CbADH. The four subunits are: A
(yellow-light green), B (red), C (blue), and D (dark green). (A)
Stereoview of the hydrogen-bond network at the interface of
three subunits of the quadruple mutant of CbADH. Residues
Asp237, Lys257, Arg238, Ser108, and Glu165 from Subunit A,
Arg304 from Subunit B, and Ala97 of subunit D (via its
main-chain carbonyl group) participate in this network. The
water molecules are shown in magenta, hydrogen bond in white
dashed lines, and distances are in Å. '2Fo-Fc' electron density
map of the quadruple mutant is contoured at 1.7  level. (B)
Extra salt bridges (magenta) in subunit A (the yellow coil) in
the Q165E/M304R mutant. level. (B)
Extra salt bridges (magenta) in subunit A (the yellow coil) in
the Q165E/M304R mutant.
|
 |
Figure 5.
Fig. 5. Two alternative conformations of Lys254 of the
quadruple mutant of CbADH. (A) In subunit A, the  -amino group
of Lys254 points towards Glu280 to form a salt bridge; however
neither hydrogen-bond connection nor electrostatic interaction
between Lys254 and Glu224 is observed. (B) In subunit D, Lys254
is equally distanced from both Glu280 and Glu224; it interacts
electrostatically with both of them and forms an additional
hydrogen bond via a water molecule with Glu224. '2Fo-Fc'
electron density map is contoured at 1.2 -amino group
of Lys254 points towards Glu280 to form a salt bridge; however
neither hydrogen-bond connection nor electrostatic interaction
between Lys254 and Glu224 is observed. (B) In subunit D, Lys254
is equally distanced from both Glu280 and Glu224; it interacts
electrostatically with both of them and forms an additional
hydrogen bond via a water molecule with Glu224. '2Fo-Fc'
electron density map is contoured at 1.2  . .
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the Protein Society:
Protein Sci
(2002,
11,
2561-2574)
copyright 2002.
|
|
| |
Figures were
selected
by the author.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.K.Williamson
(2008).
Structural and functional aspects of the MSP (PsbO) and study of its differences in thermophilic versus mesophilic organisms.
|
| |
Photosynth Res,
98,
365-389.
|
 |
|
|
|
|
 |
E.Goihberg,
O.Dym,
S.Tel-Or,
L.Shimon,
F.Frolow,
M.Peretz,
and
Y.Burstein
(2008).
Thermal stabilization of the protozoan Entamoeba histolytica alcohol dehydrogenase by a single proline substitution.
|
| |
Proteins,
72,
711-719.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.F.Mehl,
B.Demeler,
and
A.Zraikat
(2007).
A water mediated electrostatic interaction gives thermal stability to the "tail" region of the GrpE protein from E. coli.
|
| |
Protein J,
26,
239-245.
|
 |
|
|
|
|
 |
E.Goihberg,
O.Dym,
S.Tel-Or,
I.Levin,
M.Peretz,
and
Y.Burstein
(2007).
A single proline substitution is critical for the thermostabilization of Clostridium beijerinckii alcohol dehydrogenase.
|
| |
Proteins,
66,
196-204.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
W.Zhang,
E.J.Mullaney,
and
X.G.Lei
(2007).
Adopting selected hydrogen bonding and ionic interactions from Aspergillus fumigatus phytase structure improves the thermostability of Aspergillus niger PhyA phytase.
|
| |
Appl Environ Microbiol,
73,
3069-3076.
|
 |
|
|
|
|
 |
R.A.Frank,
J.V.Pratap,
X.Y.Pei,
R.N.Perham,
and
B.F.Luisi
(2005).
The molecular origins of specificity in the assembly of a multienzyme complex.
|
| |
Structure,
13,
1119-1130.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
Y.Eisenberg-Domovich,
V.P.Hytönen,
M.Wilchek,
E.A.Bayer,
M.S.Kulomaa,
and
O.Livnah
(2005).
High-resolution crystal structure of an avidin-related protein: insight into high-affinity biotin binding and protein stability.
|
| |
Acta Crystallogr D Biol Crystallogr,
61,
528-538.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.W.Partridge,
A.G.Therien,
and
C.M.Deber
(2004).
Missense mutations in transmembrane domains of proteins: phenotypic propensity of polar residues for human disease.
|
| |
Proteins,
54,
648-656.
|
 |
|
|
|
|
 |
J.K.Yano,
and
T.L.Poulos
(2003).
New understandings of thermostable and peizostable enzymes.
|
| |
Curr Opin Biotechnol,
14,
360-365.
|
 |
|
|
|
|
 |
Z.Xu,
Y.Liu,
Y.Yang,
W.Jiang,
E.Arnold,
and
J.Ding
(2003).
Crystal structure of D-Hydantoinase from Burkholderia pickettii at a resolution of 2.7 Angstroms: insights into the molecular basis of enzyme thermostability.
|
| |
J Bacteriol,
185,
4038-4049.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

