 |
PDBsum entry 2oui

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
2oui
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
D275p mutant of alcohol dehydrogenase from protozoa entamoeba histolytica
|
|
Structure:
|
 |
NADP-dependent alcohol dehydrogenase. Chain: a, b, c, d. Engineered: yes. Mutation: yes
|
|
Source:
|
 |
Entamoeba histolytica. Organism_taxid: 5759. Gene: adh1. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
1.77Å
|
R-factor:
|
0.150
|
R-free:
|
0.178
|
|
|
Authors:
|
 |
F.Frolow,L.Shimon,Y.Burstein,E.Goihberg,M.Peretz,O.Dym
|
Key ref:

|
 |
E.Goihberg
et al.
(2008).
Thermal stabilization of the protozoan Entamoeba histolytica alcohol dehydrogenase by a single proline substitution.
Proteins,
72,
711-719.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
11-Feb-07
|
Release date:
|
12-Feb-08
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P35630
(ADH1_ENTHI) -
NADP-dependent isopropanol dehydrogenase from Entamoeba histolytica (strain ATCC 30459 / HM-1:IMSS / ABRM)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
366 a.a.
360 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.1.1.80
- isopropanol dehydrogenase (NADP(+)).
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
propan-2-ol + NADP+ = acetone + NADPH + H+
|
 |
 |
 |
 |
 |
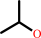 propan-2-ol
propan-2-ol
|
+
|
NADP(+)
Bound ligand (Het Group name = )
matches with 60.00% similarity
|
=
|
 acetone
acetone
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Proteins
72:711-719
(2008)
|
|
PubMed id:
|
|
|
|

|
| |
|
Thermal stabilization of the protozoan Entamoeba histolytica alcohol dehydrogenase by a single proline substitution.
|
|
E.Goihberg,
O.Dym,
S.Tel-Or,
L.Shimon,
F.Frolow,
M.Peretz,
Y.Burstein.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Analysis of the three-dimensional structures of two closely related thermophilic
and hyperthermophilic alcohol dehydrogenases (ADHs) from the respective
microorganisms Entamoeba histolytica (EhADH1) and Thermoanaerobacter brockii
(TbADH) suggested that a unique, strategically located proline residue (Pro275)
at the center of the dimerization interface might be crucial for maintaining the
thermal stability of TbADH. To assess the contribution of Pro275 to the thermal
stability of the ADHs, we applied site-directed mutagenesis to replace Asp275 of
EhADH1 with Pro (D275P-EhADH1) and conversely Pro275 of TbADH with Asp
(P275D-TbADH). The results indicate that replacing Asp275 with Pro significantly
enhances the thermal stability of EhADH1 (DeltaT(1/2) <or= +10 degrees C),
whereas the reverse mutation in the thermophilic TbADH (P275D-TbADH) reduces the
thermostability of the enzyme (DeltaT(1/2) <or= -18.8 degrees C). Analysis of
the crystal structures of the thermostabilized mutant D275P-EhADH1 and the
thermocompromised mutant P275D-TbADH suggest that a proline residue at position
275 thermostabilized the enzymes by reducing flexibility and by reinforcing
hydrophobic interactions at the dimer-dimer interface of the tetrameric ADHs.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1. Structure-based sequence alignment of ADHs from
Thermoanaerobacter brockii (TbADH, TB[18]), Clostridium
beijerinckii (CbADH, CB[18]) and Entamoeba histolytica (EhADH,
EH[19]). The secondary structure elements are shown above the
sequences with twisted rods for  -helices
and arrows for -helices
and arrows for  -strands.
The mutagenesis site (m) and the metal ligand residues (Zn) are
depicted above and below the sequence, respectively. The figure
was prepared with INDONESIA program package
http://alpha2.bmc.uu.se/ [sim.gif] -strands.
The mutagenesis site (m) and the metal ligand residues (Zn) are
depicted above and below the sequence, respectively. The figure
was prepared with INDONESIA program package
http://alpha2.bmc.uu.se/ [sim.gif] dermis/. dermis/.
|
 |
Figure 2.
Figure 2. (a) Ribbon diagram of the TbADH tetramer. Subunits
are represented in different colors. Pro residues, depicted in
ball representation, are colored magenta (Pro275) and cyan
(Pro100). The active site Zn is colored green. (b)
Superimposition of the structures of the wild-type apo-EhADH1
(colored orange) and the apo D275P-EhADH1 mutant (colored
green). Residues within a sphere of a 4-Å radius of the
mutation are shown (the superscript refers to the subunit). (c)
Superimposition of the structures of the wild-type holo-TbADH
(colored green) and the holo P275D-TbADH mutant (colored
orange). Residues within a sphere of a 4-Å radius of the
mutation are shown (the superscript refers to the subunit).
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from John Wiley & Sons, Inc.:
Proteins
(2008,
72,
711-719)
copyright 2008.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
M.K.Tiwari,
H.J.Moon,
M.Jeya,
and
J.K.Lee
(2010).
Cloning and characterization of a thermostable xylitol dehydrogenase from Rhizobium etli CFN42.
|
| |
Appl Microbiol Biotechnol,
87,
571-581.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

