 |
PDBsum entry 1ipe

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1ipe
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Tropinone reductase-ii complexed with NADPH
|
|
Structure:
|
 |
Tropinone reductase-ii. Chain: a, b. Engineered: yes
|
|
Source:
|
 |
Datura stramonium. Jimsonweed. Organism_taxid: 4076. Organ: cultured root. Expressed in: escherichia coli bl21(de3). Expression_system_taxid: 469008.
|
|
Biol. unit:
|
 |
 Tetramer (from
)
Tetramer (from
)
|
|
Resolution:
|
 |
|
2.50Å
|
R-factor:
|
0.195
|
R-free:
|
0.268
|
|
|
Authors:
|
 |
A.Yamashita,M.Endo,T.Higashi,T.Nakatsu,Y.Yamada,J.Oda,H.Kato,Riken Structural Genomics/proteomics Initiative (Rsgi)
|
Key ref:

|
 |
A.Yamashita
et al.
(2003).
Capturing enzyme structure prior to reaction initiation: tropinone reductase-II-substrate complexes.
Biochemistry,
42,
5566-5573.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
09-May-01
|
Release date:
|
03-Jun-03
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P50163
(TRN2_DATST) -
Tropinone reductase 2 from Datura stramonium
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
260 a.a.
259 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.1.1.236
- tropinone reductase Ii.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
pseudotropine + NADP+ = tropinone + NADPH + H+
|
 |
 |
 |
 |
 |
pseudotropine
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
 NADP(+)
NADP(+)
|
=
|
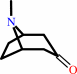 tropinone
tropinone
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
42:5566-5573
(2003)
|
|
PubMed id:
|
|
|
|

|
| |
|
Capturing enzyme structure prior to reaction initiation: tropinone reductase-II-substrate complexes.
|
|
A.Yamashita,
M.Endo,
T.Higashi,
T.Nakatsu,
Y.Yamada,
J.Oda,
H.Kato.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
To understand the catalytic mechanism of an enzyme, it is crucial to determine
the crystallographic structures corresponding to the individual reaction steps.
Here, we report two crystal structures of enzyme-substrate complexes prior to
reaction initiation: tropinone reductase-II (TR-II)-NADPH and
TR-II-NADPH-tropinone complexes, determined from the identical crystals. A
combination of two kinetic crystallographic techniques, a continuous flow of the
substrates and Laue diffraction measurements, enabled us to capture the transit
structures prior to the reaction proceeding. A structure comparison of the
enzyme-substrate complex elucidated in this study with the enzyme-product
complex in our previous study indicates that one of the substrates, tropinone,
is rotated relative to the product so as to make the spatial organization in the
active site favorable for the reaction to proceed. Side chains of the residues
in the active site also alter their conformations to keep the complementarity of
the space for the substrate or the product and to assist the rotational movement.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
M.Katzberg,
N.Skorupa-Parachin,
M.F.Gorwa-Grauslund,
and
M.Bertau
(2010).
Engineering Cofactor Preference of Ketone Reducing Biocatalysts: A Mutagenesis Study on a gamma-Diketone Reductase from the Yeast Saccharomyces cerevisiae Serving as an Example.
|
| |
Int J Mol Sci,
11,
1735-1758.
|
 |
|
|
|
|
 |
A.Brock,
W.Brandt,
and
B.Dräger
(2008).
The functional divergence of short-chain dehydrogenases involved in tropinone reduction.
|
| |
Plant J,
54,
388-401.
|
 |
|
|
|
|
 |
A.C.Freydank,
W.Brandt,
and
B.Dräger
(2008).
Protein structure modeling indicates hexahistidine-tag interference with enzyme activity.
|
| |
Proteins,
72,
173-183.
|
 |
|
|
|
|
 |
A.T.Keatinge-Clay
(2007).
A tylosin ketoreductase reveals how chirality is determined in polyketides.
|
| |
Chem Biol,
14,
898-908.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.Stöckigt,
and
S.Panjikar
(2007).
Structural biology in plant natural product biosynthesis--architecture of enzymes from monoterpenoid indole and tropane alkaloid biosynthesis.
|
| |
Nat Prod Rep,
24,
1382-1400.
|
 |
|
|
|
|
 |
D.Bourgeois,
and
A.Royant
(2005).
Advances in kinetic protein crystallography.
|
| |
Curr Opin Struct Biol,
15,
538-547.
|
 |
|
|
|
|
 |
Q.S.Hanley,
and
M.B.Denton
(2005).
Advances in array detectors for X-ray diffraction techniques.
|
| |
J Synchrotron Radiat,
12,
618-625.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

