 |
PDBsum entry 1h3b

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Isomerase
|
 |
|
Title:
|
 |
Squalene-hopene cyclase
|
|
Structure:
|
 |
Squalene--hopene cyclase. Chain: a, b, c. Synonym: shc. Engineered: yes
|
|
Source:
|
 |
Alicyclobacillus acidocaldarius. Organism_taxid: 405212. Expressed in: escherichia coli k-12. Expression_system_taxid: 83333. Expression_system_variant: jm105. Other_details: thermostable, acidophilic
|
|
Biol. unit:
|
 |
Dimer (from PDB file)
|
|
Resolution:
|
 |
|
2.80Å
|
R-factor:
|
0.206
|
R-free:
|
0.250
|
|
|
Authors:
|
 |
A.Lenhart,D.J.Reinert,W.A.Weihofen,J.D.Aebi,H.Dehmlow,O.H.Morand, G.E.Schulz
|
|
Key ref:
|
 |
A.Lenhart
et al.
(2003).
Binding structures and potencies of oxidosqualene cyclase inhibitors with the homologous squalene-hopene cyclase.
J Med Chem,
46,
2083-2092.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
25-Aug-02
|
Release date:
|
21-Aug-03
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P33247
(SQHC_ALIAD) -
Squalene--hopene cyclase from Alicyclobacillus acidocaldarius subsp. acidocaldarius (strain ATCC 27009 / DSM 446 / BCRC 14685 / JCM 5260 / KCTC 1825 / NBRC 15652 / NCIMB 11725 / NRRL B-14509 / 104-IA)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
631 a.a.
620 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.4.2.1.129
- squalene--hopanol cyclase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
squalene + H2O = hopan-22-ol
|
 |
 |
 |
 |
 |
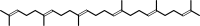 squalene
squalene
|
+
|
H2O
|
=
|
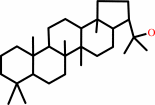 hopan-22-ol
hopan-22-ol
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.5.4.99.17
- squalene--hopene cyclase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
|
 |
 |
 |
 |
 |
Reaction:
|
 |
squalene = hop-2229-ene
|
 |
 |
 |
 |
 |
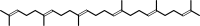 squalene
squalene
|
=
|
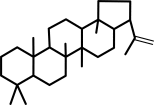 hop-22(29)-ene
hop-22(29)-ene
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Med Chem
46:2083-2092
(2003)
|
|
PubMed id:
|
|
|
|

|
| |
|
Binding structures and potencies of oxidosqualene cyclase inhibitors with the homologous squalene-hopene cyclase.
|
|
A.Lenhart,
D.J.Reinert,
J.D.Aebi,
H.Dehmlow,
O.H.Morand,
G.E.Schulz.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The binding structures of 11 human oxidosqualene cyclase inhibitors designed as
cholesterol-lowering agents were determined for the squalene-hopene cyclase from
Alicyclobacillus acidocaldarius, which is the only structurally known homologue
of the human enzyme. The complexes were produced by cocrystallization, and the
structures were elucidated by X-ray diffraction analyses. All inhibitors were
bound in the large active center cavity. The detailed binding structures are
presented and discussed in the light of the IC50 values of these 11 as well as
17 other inhibitors. They provide a consistent picture for the inhibition of the
bacterial enzyme and can be used to adjust and improve homology models of the
human enzyme. The detailed active center structures of the two enzymes are too
different to show an IC50 correlation.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
S.Z.Grinter,
Y.Liang,
S.Y.Huang,
S.M.Hyder,
and
X.Zou
(2011).
An inverse docking approach for identifying new potential anti-cancer targets.
|
| |
J Mol Graph Model,
29,
795-799.
|
 |
|
|
|
|
 |
R.J.Cenedella
(2009).
Cholesterol synthesis inhibitor U18666A and the role of sterol metabolism and trafficking in numerous pathophysiological processes.
|
| |
Lipids,
44,
477-487.
|
 |
|
|
|
|
 |
A.N.Eddine,
J.P.von Kries,
M.V.Podust,
T.Warrier,
S.H.Kaufmann,
and
L.M.Podust
(2008).
X-ray structure of 4,4'-dihydroxybenzophenone mimicking sterol substrate in the active site of sterol 14alpha-demethylase (CYP51).
|
| |
J Biol Chem,
283,
15152-15159.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
L.M.Podust,
J.P.von Kries,
A.N.Eddine,
Y.Kim,
L.V.Yermalitskaya,
R.Kuehne,
H.Ouellet,
T.Warrier,
M.Alteköster,
J.S.Lee,
J.Rademann,
H.Oschkinat,
S.H.Kaufmann,
and
M.R.Waterman
(2007).
Small-molecule scaffolds for CYP51 inhibitors identified by high-throughput screening and defined by X-ray crystallography.
|
| |
Antimicrob Agents Chemother,
51,
3915-3923.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Oliaro-Bosso,
F.Viola,
S.Taramino,
S.Tagliapietra,
A.Barge,
G.Cravotto,
and
G.Balliano
(2007).
Inhibitory Effect of Umbelliferone Aminoalkyl Derivatives on Oxidosqualene Cyclases from S. cerevisiae, T. cruzi, P. carinii, H. sapiens, and A. thaliana: a Structure-Activity Study.
|
| |
ChemMedChem,
2,
226-233.
|
 |
|
|
|
|
 |
K.Schärer,
M.Morgenthaler,
R.Paulini,
U.Obst-Sander,
D.W.Banner,
D.Schlatter,
J.Benz,
M.Stihle,
and
F.Diederich
(2005).
Quantification of cation-pi interactions in protein-ligand complexes: crystal-structure analysis of Factor Xa bound to a quaternary ammonium ion ligand.
|
| |
Angew Chem Int Ed Engl,
44,
4400-4404.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
K.U.Wendt
(2005).
Enzyme mechanisms for triterpene cyclization: new pieces of the puzzle.
|
| |
Angew Chem Int Ed Engl,
44,
3966-3971.
|
 |
|
|
|
|
 |
R.J.Cenedella,
P.S.Sexton,
K.Krishnan,
and
D.F.Covey
(2005).
Comparison of effects of U18666A and enantiomeric U18666A on sterol synthesis and induction of apoptosis.
|
| |
Lipids,
40,
635-640.
|
 |
|
|
|
|
 |
S.Oliaro-Bosso,
T.Schulz-Gasch,
G.Balliano,
and
F.Viola
(2005).
Access of the substrate to the active site of yeast oxidosqualene cyclase: an inhibition and site-directed mutagenesis approach.
|
| |
Chembiochem,
6,
2221-2228.
|
 |
|
|
|
|
 |
D.J.Reinert,
G.Balliano,
and
G.E.Schulz
(2004).
Conversion of squalene to the pentacarbocyclic hopene.
|
| |
Chem Biol,
11,
121-126.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.Oliaro-Bosso,
F.Viola,
S.Matsuda,
G.Cravotto,
S.Tagliapietra,
and
G.Balliano
(2004).
Umbelliferone aminoalkyl derivatives as inhibitors of oxidosqualene cyclases from Saccharomyces cerevisiae, Trypanosoma cruzi, and Pneumocystis carinii.
|
| |
Lipids,
39,
1007-1012.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

