 |
PDBsum entry 1f12

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1f12
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.1.1.35
- 3-hydroxyacyl-CoA dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a (3S)-3-hydroxyacyl-CoA + NAD+ = a 3-oxoacyl-CoA + NADH + H+
|
 |
 |
 |
 |
 |
(3S)-3-hydroxyacyl-CoA
Bound ligand (Het Group name = )
matches with 96.36% similarity
|
+
|
 NAD(+)
NAD(+)
|
=
|
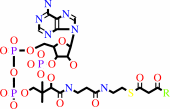 3-oxoacyl-CoA
3-oxoacyl-CoA
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
275:27186-27196
(2000)
|
|
PubMed id:
|
|
|
|

|
| |
|
Sequestration of the active site by interdomain shifting. Crystallographic and spectroscopic evidence for distinct conformations of L-3-hydroxyacyl-CoA dehydrogenase.
|
|
J.J.Barycki,
L.K.O'Brien,
A.W.Strauss,
L.J.Banaszak.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
l-3-Hydroxyacyl-CoA dehydrogenase reversibly catalyzes the conversion of
l-3-hydroxyacyl-CoA to 3-ketoacyl-CoA concomitant with the reduction of NAD(+)
to NADH as part of the beta-oxidation spiral. In this report, crystal structures
have been solved for the apoenzyme, binary complexes of the enzyme with reduced
cofactor or 3-hydroxybutyryl-CoA substrate, and an abortive ternary complex of
the enzyme with NAD(+) and acetoacetyl-CoA. The models illustrate positioning of
cofactor and substrate within the active site of the enzyme. Comparison of these
structures with the previous model of the enzyme-NAD(+) complex reveals that
although significant shifting of the NAD(+)-binding domain relative to the
C-terminal domain occurs in the ternary and substrate-bound complexes, there are
few differences between the apoenzyme and cofactor-bound complexes. Analysis of
these models clarifies the role of key amino acids implicated in catalysis and
highlights additional critical residues. Furthermore, a novel charge transfer
complex has been identified in the course of abortive ternary complex formation,
and its characterization provides additional insight into aspects of the
catalytic mechanism of l-3-hydroxyacyl-CoA dehydrogenase.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 2.
Fig. 2. L-3-Hydroxyacyl-CoA dehydrogenase complexed with
the substrate, 3-hydroxybutyryl-CoA. The ribbon diagram depicts
the two-domain structure of an HAD subunit, with the first 200
amino acids comprising the NAD +-binding domain and the
remaining residues comprising the C-terminal domain.
3-Hydroxybutyryl-CoA, shown in ball and stick representation,
binds within the cleft between these two domains. The adenine
moiety of coenzyme A is positioned adjacent to the
helix-turn-helix tail (  2- 2-  3) of the
NAD^+-binding domain, and the acyl chain is within the enzyme
active site. 3) of the
NAD^+-binding domain, and the acyl chain is within the enzyme
active site.
|
 |
Figure 4.
Fig. 4. Schematic of the 3-hydroxybutyryl-CoA-binding
site. Hydrogen bonds involved in binding of the substrate,
3-hydroxybutyryl-CoA, to the apoenzyme are represented as dashed
lines, with residues of the opposing subunit of the dimer
indicated with an asterisk.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2000,
275,
27186-27196)
copyright 2000.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
J.Arima,
A.Uesumi,
H.Mitsuzumi,
and
N.Mori
(2010).
Biochemical characterization of L-carnitine dehydrogenases from Rhizobium sp. and Xanthomonas translucens.
|
| |
Biosci Biotechnol Biochem,
74,
1237-1242.
|
 |
|
|
|
|
 |
J.Parkot,
H.Gröger,
and
W.Hummel
(2010).
Purification, cloning, and overexpression of an alcohol dehydrogenase from Nocardia globerula reducing aliphatic ketones and bulky ketoesters.
|
| |
Appl Microbiol Biotechnol,
86,
1813-1820.
|
 |
|
|
|
|
 |
Y.Asada,
C.Kuroishi,
Y.Ukita,
R.Sumii,
S.Endo,
T.Matsunaga,
A.Hara,
and
N.Kunishima
(2008).
Crystallization and preliminary X-ray crystallographic analysis of rabbit L-gulonate 3-dehydrogenase.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
64,
228-230.
|
 |
|
|
|
|
 |
A.Li,
T.Itoh,
T.Taguchi,
T.Xiang,
Y.Ebizuka,
and
K.Ichinose
(2005).
Functional studies on a ketoreductase gene from Streptomyces sp. AM-7161 to control the stereochemistry in medermycin biosynthesis.
|
| |
Bioorg Med Chem,
13,
6856-6863.
|
 |
|
|
|
|
 |
S.Y.Yang,
X.Y.He,
and
H.Schulz
(2005).
3-Hydroxyacyl-CoA dehydrogenase and short chain 3-hydroxyacyl-CoA dehydrogenase in human health and disease.
|
| |
FEBS J,
272,
4874-4883.
|
 |
|
|
|
|
 |
M.Ishikawa,
D.Tsuchiya,
T.Oyama,
Y.Tsunaka,
and
K.Morikawa
(2004).
Structural basis for channelling mechanism of a fatty acid beta-oxidation multienzyme complex.
|
| |
EMBO J,
23,
2745-2754.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.A.Bottoms,
P.E.Smith,
and
J.J.Tanner
(2002).
A structurally conserved water molecule in Rossmann dinucleotide-binding domains.
|
| |
Protein Sci,
11,
2125-2137.
|
 |
|
|
|
|
 |
X.Y.He,
G.Merz,
Y.Z.Yang,
P.Mehta,
H.Schulz,
and
S.Y.Yang
(2001).
Characterization and localization of human type10 17beta-hydroxysteroid dehydrogenase.
|
| |
Eur J Biochem,
268,
4899-4907.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

