 |
PDBsum entry 1es3

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.4.16.4
- serine-type D-Ala-D-Ala carboxypeptidase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
D-alanyl-D-alanine + H2O = 2 D-alanine
|
 |
 |
 |
 |
 |
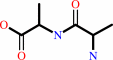
|
+
|
|
=
|
 2
×
2
×
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
42:2895-2906
(2003)
|
|
PubMed id:
|
|
|
|

|
| |
|
Catalytic mechanism of the Streptomyces K15 DD-transpeptidase/penicillin-binding protein probed by site-directed mutagenesis and structural analysis.
|
|
N.Rhazi,
P.Charlier,
D.Dehareng,
D.Engher,
M.Vermeire,
J.M.Frère,
M.Nguyen-Distèche,
E.Fonzé.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The Streptomyces K15 penicillin-binding DD-transpeptidase is presumed to be
involved in peptide cross-linking during bacterial cell wall peptidoglycan
assembly. To gain insight into the catalytic mechanism, the roles of residues
Lys38, Ser96, and Cys98, belonging to the structural elements defining the
active site cleft, have been investigated by site-directed mutagenesis,
biochemical studies, and X-ray diffraction analysis. The Lys38His and Ser96Ala
mutations almost completely abolished the penicillin binding and severely
impaired the transpeptidase activities while the geometry of the active site was
essentially the same as in the wild-type enzyme. It is proposed that Lys38 acts
as the catalytic base that abstracts a proton from the active serine Ser35
during nucleophilic attack and that Ser96 is a key intermediate in the proton
transfer from the Ogamma of Ser35 to the substrate leaving group nitrogen. The
role of these two residues should be conserved among penicillin-binding proteins
containing the Ser-Xaa-Asn/Cys sequence in motif 2. Conversion of Cys98 into Asn
decreased the transpeptidase activity and increased hydrolysis of the thiolester
substrate and the acylation rate with most beta-lactam antibiotics. Cys98 is
proposed to play the same role as Asn in motif 2 of other penicilloyl serine
transferases in properly positioning the substrate for the catalytic process.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
C.Urbach,
J.Fastrez,
and
P.Soumillion
(2008).
A New Family of Cyanobacterial Penicillin-binding Proteins: A MISSING LINK IN THE EVOLUTION OF CLASS A {beta}-LACTAMASES.
|
| |
J Biol Chem,
283,
32516-32526.
|
 |
|
|
|
|
 |
P.Macheboeuf,
C.Contreras-Martel,
V.Job,
O.Dideberg,
and
A.Dessen
(2006).
Penicillin binding proteins: key players in bacterial cell cycle and drug resistance processes.
|
| |
FEMS Microbiol Rev,
30,
673-691.
|
 |
|
|
|
|
 |
M.E.Stefanova,
J.Tomberg,
C.Davies,
R.A.Nicholas,
and
W.G.Gutheil
(2004).
Overexpression and enzymatic characterization of Neisseria gonorrhoeae penicillin-binding protein 4.
|
| |
Eur J Biochem,
271,
23-32.
|
 |
|
|
|
|
 |
M.S.Wilke,
T.L.Hills,
H.Z.Zhang,
H.F.Chambers,
and
N.C.Strynadka
(2004).
Crystal structures of the Apo and penicillin-acylated forms of the BlaR1 beta-lactam sensor of Staphylococcus aureus.
|
| |
J Biol Chem,
279,
47278-47287.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Y.Zong,
S.K.Mazmanian,
O.Schneewind,
and
S.V.Narayana
(2004).
The structure of sortase B, a cysteine transpeptidase that tethers surface protein to the Staphylococcus aureus cell wall.
|
| |
Structure,
12,
105-112.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

