 |
PDBsum entry 1dch

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Transcriptional stimulator,dimerization
|
PDB id
|
|

|

|
1dch
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transcriptional stimulator,dimerization
|
 |
|
Title:
|
 |
Crystal structure of dcoh, a bifunctional, protein-binding transcription coactivator
|
|
Structure:
|
 |
Dcoh (dimerization cofactor of hnf-1). Chain: a, b, c, d, e, f, g, h. Synonym: phs, phenylalanine hydroxylase stimulator protein, 4a- carbinolamine dehydratase, pcd. Engineered: yes
|
|
Source:
|
 |
Rattus norvegicus. Norway rat. Organism_taxid: 10116. Organ: liver. Expressed in: escherichia coli. Expression_system_taxid: 562. Other_details: gst-fusion
|
|
Biol. unit:
|
 |
 Tetramer (from
)
Tetramer (from
)
|
|
Resolution:
|
 |
|
|
Authors:
|
 |
J.A.Endrizzi,J.D.Cronk,W.Wang,G.R.Crabtree,T.Alber
|
|
Key ref:
|
 |
J.A.Endrizzi
et al.
(1995).
Crystal structure of DCoH, a bifunctional, protein-binding transcriptional coactivator.
Science,
268,
556-559.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
24-Jan-95
|
Release date:
|
08-Mar-96
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P61459
(PHS_RAT) -
Pterin-4-alpha-carbinolamine dehydratase from Rattus norvegicus
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
104 a.a.
99 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.2.1.96
- 4a-hydroxytetrahydrobiopterin dehydratase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Biopterin Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(4aS,6R)-4a-hydroxy-L-erythro-5,6,7,8-tetrahydrobiopterin = (6R)-L- erythro-6,7-dihydrobiopterin + H2O
|
 |
 |
 |
 |
 |
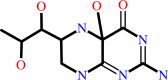 4a-hydroxytetrahydrobiopterin
4a-hydroxytetrahydrobiopterin
|
=
|
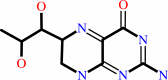 6,7-dihydrobiopterin
6,7-dihydrobiopterin
|
+
|
H(2)O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Science
268:556-559
(1995)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of DCoH, a bifunctional, protein-binding transcriptional coactivator.
|
|
J.A.Endrizzi,
J.D.Cronk,
W.Wang,
G.R.Crabtree,
T.Alber.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
DCoH, the dimerization cofactor of hepatocyte nuclear factor-1, stimulates gene
expression by associating with specific DNA binding proteins and also catalyzes
the dehydration of the biopterin cofactor of phenylalanine hydroxylase. The
x-ray crystal structure determined at 3 angstrom resolution reveals that DCoH
forms a tetramer containing two saddle-shaped grooves that comprise likely
macromolecule binding sites. Two equivalent enzyme active sites flank each
saddle, suggesting that there is a spatial connection between the catalytic and
binding activities. Structural similarities between the DCoH fold and nucleic
acid-binding proteins argue that the saddle motif has evolved to bind diverse
ligands or that DCoH unexpectedly may bind nucleic acids.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
T.Jin,
F.Guo,
I.G.Serebriiskii,
A.Howard,
and
Y.Z.Zhang
(2007).
A 1.55 A resolution X-ray crystal structure of HEF2/ERH and insights into its transcriptional and cell-cycle interaction networks.
|
| |
Proteins,
68,
427-437.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
E.P.von Strandmann,
S.Senkel,
G.Ryffel,
and
U.R.Hengge
(2001).
Dimerization co-factor of hepatocyte nuclear factor 1/pterin-4alpha-carbinolamine dehydratase is necessary for pigmentation in Xenopus and overexpressed in primary human melanoma lesions.
|
| |
Am J Pathol,
158,
2021-2029.
|
 |
|
|
|
|
 |
E.Pogge von Strandmann,
S.Senkel,
and
G.U.Ryffel
(2001).
ERH (enhancer of rudimentary homologue), a conserved factor identical between frog and human, is a transcriptional repressor.
|
| |
Biol Chem,
382,
1379-1385.
|
 |
|
|
|
|
 |
Q.X.Hua,
M.Zhao,
N.Narayana,
S.H.Nakagawa,
W.Jia,
and
M.A.Weiss
(2000).
Diabetes-associated mutations in a beta-cell transcription factor destabilize an antiparallel "mini-zipper" in a dimerization interface.
|
| |
Proc Natl Acad Sci U S A,
97,
1999-2004.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.Song,
T.Xia,
and
R.A.Jensen
(1999).
PhhB, a Pseudomonas aeruginosa homolog of mammalian pterin 4a-carbinolamine dehydratase/DCoH, does not regulate expression of phenylalanine hydroxylase at the transcriptional level.
|
| |
J Bacteriol,
181,
2789-2796.
|
 |
|
|
|
|
 |
X.D.Lei,
and
S.Kaufman
(1999).
Characterization of expression of the gene for human pterin carbinolamine dehydratase/dimerization cofactor of HNF1.
|
| |
DNA Cell Biol,
18,
243-252.
|
 |
|
|
|
|
 |
A.K.Aggarwal,
and
D.A.Wah
(1998).
Novel site-specific DNA endonucleases.
|
| |
Curr Opin Struct Biol,
8,
19-25.
|
 |
|
|
|
|
 |
I.Rebrin,
B.Thöny,
S.W.Bailey,
and
J.E.Ayling
(1998).
Stereospecificity and catalytic function of histidine residues in 4a-hydroxy-tetrahydropterin dehydratase/DCoH.
|
| |
Biochemistry,
37,
11246-11254.
|
 |
|
|
|
|
 |
S.Köster,
G.Stier,
N.Kubasch,
H.C.Curtius,
and
S.Ghisla
(1998).
Pterin-4a-carbinolamine dehydratase from Pseudomonas aeruginosa: characterization, catalytic mechanism and comparison to the human enzyme.
|
| |
Biol Chem,
379,
1427-1432.
|
 |
|
|
|
|
 |
X.D.Lei,
and
S.Kaufman
(1998).
Identification of hepatic nuclear factor 1 binding sites in the 5' flanking region of the human phenylalanine hydroxylase gene: implication of a dual function of phenylalanine hydroxylase stimulator in the phenylalanine hydroxylation system.
|
| |
Proc Natl Acad Sci U S A,
95,
1500-1504.
|
 |
|
|
|
|
 |
A.K.Aggarwal
(1997).
Homing in on intron-encoded endonucleases.
|
| |
Nat Struct Biol,
4,
423-424.
|
 |
|
|
|
|
 |
A.V.Efimov
(1997).
Structural trees for protein superfamilies.
|
| |
Proteins,
28,
241-260.
|
 |
|
|
|
|
 |
D.E.Birse,
U.Kapp,
K.Strub,
S.Cusack,
and
A.Aberg
(1997).
The crystal structure of the signal recognition particle Alu RNA binding heterodimer, SRP9/14.
|
| |
EMBO J,
16,
3757-3766.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
D.J.Sourdive,
C.Transy,
S.Garbay,
and
M.Yaniv
(1997).
The bifunctional DCOH protein binds to HNF1 independently of its 4-alpha-carbinolamine dehydratase activity.
|
| |
Nucleic Acids Res,
25,
1476-1484.
|
 |
|
|
|
|
 |
G.Auerbach,
A.Herrmann,
M.Gütlich,
M.Fischer,
U.Jacob,
A.Bacher,
and
R.Huber
(1997).
The 1.25 A crystal structure of sepiapterin reductase reveals its binding mode to pterins and brain neurotransmitters.
|
| |
EMBO J,
16,
7219-7230.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
G.Johnen,
and
S.Kaufman
(1997).
Studies on the enzymatic and transcriptional activity of the dimerization cofactor for hepatocyte nuclear factor 1.
|
| |
Proc Natl Acad Sci U S A,
94,
13469-13474.
|
 |
|
|
|
|
 |
P.J.Heath,
K.M.Stephens,
R.J.Monnat,
and
B.L.Stoddard
(1997).
The structure of I-Crel, a group I intron-encoded homing endonuclease.
|
| |
Nat Struct Biol,
4,
468-476.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.G.Murzin
(1996).
Structural classification of proteins: new superfamilies.
|
| |
Curr Opin Struct Biol,
6,
386-394.
|
 |
|
|
|
|
 |
G.R.Crabtree,
and
S.L.Schreiber
(1996).
Three-part inventions: intracellular signaling and induced proximity.
|
| |
Trends Biochem Sci,
21,
418-422.
|
 |
|
|
|
|
 |
J.D.Cronk,
J.A.Endrizzi,
and
T.Alber
(1996).
High-resolution structures of the bifunctional enzyme and transcriptional coactivator DCoH and its complex with a product analogue.
|
| |
Protein Sci,
5,
1963-1972.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
N.C.Strynadka,
S.E.Jensen,
P.M.Alzari,
and
M.N.James
(1996).
A potent new mode of beta-lactamase inhibition revealed by the 1.7 A X-ray crystallographic structure of the TEM-1-BLIP complex.
|
| |
Nat Struct Biol,
3,
290-297.
|
 |
|
|
|
|
 |
S.Köster,
G.Stier,
R.Ficner,
M.Hölzer,
H.C.Curtius,
D.Suck,
and
S.Ghisla
(1996).
Location of the active site and proposed catalytic mechanism of pterin-4a-carbinolamine dehydratase.
|
| |
Eur J Biochem,
241,
858-864.
|
 |
|
|
|
|
 |
S.K.Burley
(1996).
The TATA box binding protein.
|
| |
Curr Opin Struct Biol,
6,
69-75.
|
 |
|
|
|
|
 |
G.Johnen,
D.Kowlessur,
B.A.Citron,
and
S.Kaufman
(1995).
Characterization of the wild-type form of 4a-carbinolamine dehydratase and two naturally occurring mutants associated with hyperphenylalaninemia.
|
| |
Proc Natl Acad Sci U S A,
92,
12384-12388.
|
 |
|
|
|
|
 |
R.Kanaar,
A.L.Lee,
D.Z.Rudner,
D.E.Wemmer,
and
D.C.Rio
(1995).
Interaction of the sex-lethal RNA binding domains with RNA.
|
| |
EMBO J,
14,
4530-4539.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

