
|

|

|
|
 |
Contents |
 |
|
|

|

|

|

|

|

|

|
 189 a.a.
189 a.a.
|
 |

|

|

|

|

|

|

|
 190 a.a.
190 a.a.
|
 |

|

|

|

|

|

|

|
 329 a.a.
329 a.a.
|
 |

|

|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Lyase/lyase/signaling protein
|
 |
|
Title:
|
 |
Complex of gs-alpha with the catalytic domains of mammalian adenylyl cyclase: complex with 2'-deoxy-adenosine 3'-monophosphate, pyrophosphate and mg
|
|
Structure:
|
 |
Type v adenylate cyclase. Chain: a. Fragment: c1a domain. Synonym: atp pyrophosphate-lyase. Engineered: yes. Mutation: yes. Type ii adenylate cyclase. Chain: b. Fragment: c2a domain.
|
|
Source:
|
 |
Canis lupus familiaris. Dog. Organism_taxid: 9615. Strain: familiaris. Organ: heart. Expressed in: escherichia coli. Expression_system_taxid: 562. Rattus norvegicus. Norway rat.
|
|
Biol. unit:
|
 |
Trimer (from
)
|
|
Resolution:
|
 |
|
2.50Å
|
R-factor:
|
0.216
|
R-free:
|
0.264
|
|
|
Authors:
|
 |
J.J.G.Tesmer,C.A.Dessauer,R.K.Sunahara,R.A.Johnson,A.G.Gilman, S.R.Sprang
|
Key ref:

|
 |
J.J.Tesmer
et al.
(2000).
Molecular basis for P-site inhibition of adenylyl cyclase.
Biochemistry,
39,
14464-14471.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
16-Aug-99
|
Release date:
|
10-Jan-01
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P30803
(ADCY5_CANLF) -
Adenylate cyclase type 5 from Canis lupus familiaris
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
1265 a.a.
189 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
Chains A, B:
E.C.4.6.1.1
- adenylate cyclase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
ATP = 3',5'-cyclic AMP + diphosphate
|
 |
 |
 |
 |
 |
ATP
Bound ligand (Het Group name = )
matches with 90.91% similarity
|
=
|
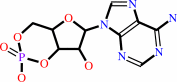 3',5'-cyclic AMP
3',5'-cyclic AMP
|
+
|
diphosphate
Bound ligand (Het Group name = )
corresponds exactly
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
Pyridoxal 5'-phosphate
Bound ligand (Het Group name =
101)
matches with 46.15% similarity
|
|
 |
 |
Enzyme class 2:
|
 |
Chain C:
E.C.3.6.5.-
- ?????
|
|
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
39:14464-14471
(2000)
|
|
PubMed id:
|
|
|
|

|
| |
|
Molecular basis for P-site inhibition of adenylyl cyclase.
|
|
J.J.Tesmer,
C.W.Dessauer,
R.K.Sunahara,
L.D.Murray,
R.A.Johnson,
A.G.Gilman,
S.R.Sprang.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
P-site inhibitors are adenosine and adenine nucleotide analogues that inhibit
adenylyl cyclase, the effector enzyme that catalyzes the synthesis of cyclic AMP
from ATP. Some of these inhibitors may represent physiological regulators of
adenylyl cyclase, and the most potent may ultimately serve as useful therapeutic
agents. Described here are crystal structures of the catalytic core of adenylyl
cyclase complexed with two such P-site inhibitors, 2'-deoxyadenosine
3'-monophosphate (2'-d-3'-AMP) and 2',5'-dideoxyadenosine 3'-triphosphate
(2',5'-dd-3'-ATP). Both inhibitors bind in the active site yet exhibit non- or
uncompetitive patterns of inhibition. While most P-site inhibitors require
pyrophosphate (PP(i)) as a coinhibitor, 2',5'-dd-3'-ATP is a potent inhibitor by
itself. The crystal structure reveals that this inhibitor exhibits two binding
modes: one with the nucleoside moiety bound to the nucleoside binding pocket of
the enzyme and the other with the beta and gamma phosphates bound to the
pyrophosphate site of the 2'-d-3'-AMP.PP(i) complex. A single metal binding site
is observed in the complex with 2'-d-3'-AMP, whereas two are observed in the
complex with 2', 5'-dd-3'-ATP. Even though P-site inhibitors are typically 10
times more potent in the presence of Mn(2+), the electron density maps reveal no
inherent preference of either metal site for Mn(2+) over Mg(2+). 2',5'-dd-3'-ATP
binds to the catalytic core of adenylyl cyclase with a K(d) of 2.4 microM in the
presence of Mg(2+) and 0.2 microM in the presence of Mn(2+). Pyrophosphate does
not compete with 2',5'-dd-3'-ATP and enhances inhibition.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
B.Pavan,
C.Biondi,
and
A.Dalpiaz
(2009).
Adenylyl cyclases as innovative therapeutic goals.
|
| |
Drug Discov Today,
14,
982-991.
|
 |
|
|
|
|
 |
R.Sadana,
and
C.W.Dessauer
(2009).
Physiological roles for G protein-regulated adenylyl cyclase isoforms: insights from knockout and overexpression studies.
|
| |
Neurosignals,
17,
5.
|
 |
|
|
|
|
 |
S.Pierre,
T.Eschenhagen,
G.Geisslinger,
and
K.Scholich
(2009).
Capturing adenylyl cyclases as potential drug targets.
|
| |
Nat Rev Drug Discov,
8,
321-335.
|
 |
|
|
|
|
 |
S.Suryanarayana,
M.Göttle,
M.Hübner,
A.Gille,
T.C.Mou,
S.R.Sprang,
M.Richter,
and
R.Seifert
(2009).
Differential inhibition of various adenylyl cyclase isoforms and soluble guanylyl cyclase by 2',3'-O-(2,4,6-trinitrophenyl)-substituted nucleoside 5'-triphosphates.
|
| |
J Pharmacol Exp Ther,
330,
687-695.
|
 |
|
|
|
|
 |
T.C.Mou,
N.Masada,
D.M.Cooper,
and
S.R.Sprang
(2009).
Structural basis for inhibition of mammalian adenylyl cyclase by calcium.
|
| |
Biochemistry,
48,
3387-3397.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.Schlicker,
A.Rauch,
K.C.Hess,
B.Kachholz,
L.R.Levin,
J.Buck,
and
C.Steegborn
(2008).
Structure-based development of novel adenylyl cyclase inhibitors.
|
| |
J Med Chem,
51,
4456-4464.
|
 |
|
|
|
|
 |
M.Nakachi,
M.Matsumoto,
P.M.Terry,
R.L.Cerny,
and
H.Moriyama
(2008).
Identification of guanylate cyclases and related signaling proteins in sperm tail from sea stars by mass spectrometry.
|
| |
Mar Biotechnol (NY),
10,
564-571.
|
 |
|
|
|
|
 |
J.L.Wang,
J.X.Guo,
Q.Y.Zhang,
J.J.Wu,
R.Seifert,
and
G.H.Lushington
(2007).
A conformational transition in the adenylyl cyclase catalytic site yields different binding modes for ribosyl-modified and unmodified nucleotide inhibitors.
|
| |
Bioorg Med Chem,
15,
2993-3002.
|
 |
|
|
|
|
 |
S.Diel,
K.Klass,
B.Wittig,
and
C.Kleuss
(2006).
Gbetagamma activation site in adenylyl cyclase type II. Adenylyl cyclase type III is inhibited by Gbetagamma.
|
| |
J Biol Chem,
281,
288-294.
|
 |
|
|
|
|
 |
I.Tews,
F.Findeisen,
I.Sinning,
A.Schultz,
J.E.Schultz,
and
J.U.Linder
(2005).
The structure of a pH-sensing mycobacterial adenylyl cyclase holoenzyme.
|
| |
Science,
308,
1020-1023.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.C.Sinha,
M.Wetterer,
S.R.Sprang,
J.E.Schultz,
and
J.U.Linder
(2005).
Origin of asymmetry in adenylyl cyclases: structures of Mycobacterium tuberculosis Rv1900c.
|
| |
EMBO J,
24,
663-673.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
T.C.Mou,
A.Gille,
D.A.Fancy,
R.Seifert,
and
S.R.Sprang
(2005).
Structural basis for the inhibition of mammalian membrane adenylyl cyclase by 2 '(3')-O-(N-Methylanthraniloyl)-guanosine 5 '-triphosphate.
|
| |
J Biol Chem,
280,
7253-7261.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.Gille,
G.H.Lushington,
T.C.Mou,
M.B.Doughty,
R.A.Johnson,
and
R.Seifert
(2004).
Differential inhibition of adenylyl cyclase isoforms and soluble guanylyl cyclase by purine and pyrimidine nucleotides.
|
| |
J Biol Chem,
279,
19955-19969.
|
 |
|
|
|
|
 |
K.Iwatsubo,
S.Minamisawa,
T.Tsunematsu,
M.Nakagome,
Y.Toya,
J.E.Tomlinson,
S.Umemura,
R.M.Scarborough,
D.E.Levy,
and
Y.Ishikawa
(2004).
Direct inhibition of type 5 adenylyl cyclase prevents myocardial apoptosis without functional deterioration.
|
| |
J Biol Chem,
279,
40938-40945.
|
 |
|
|
|
|
 |
M.Spehr,
K.Schwane,
J.A.Riffell,
J.Barbour,
R.K.Zimmer,
E.M.Neuhaus,
and
H.Hatt
(2004).
Particulate adenylate cyclase plays a key role in human sperm olfactory receptor-mediated chemotaxis.
|
| |
J Biol Chem,
279,
40194-40203.
|
 |
|
|
|
|
 |
Q.Guo,
Y.Shen,
N.L.Zhukovskaya,
J.Florián,
and
W.J.Tang
(2004).
Structural and kinetic analyses of the interaction of anthrax adenylyl cyclase toxin with reaction products cAMP and pyrophosphate.
|
| |
J Biol Chem,
279,
29427-29435.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.C.Pierre,
J.Häusler,
K.Birod,
G.Geisslinger,
and
K.Scholich
(2004).
PAM mediates sustained inhibition of cAMP signaling by sphingosine-1-phosphate.
|
| |
EMBO J,
23,
3031-3040.
|
 |
|
|
|
|
 |
K.Iwatsubo,
T.Tsunematsu,
and
Y.Ishikawa
(2003).
Isoform-specific regulation of adenylyl cyclase: a potential target in future pharmacotherapy.
|
| |
Expert Opin Ther Targets,
7,
441-451.
|
 |
|
|
|
|
 |
S.Soelaiman,
B.Q.Wei,
P.Bergson,
Y.S.Lee,
Y.Shen,
M.Mrksich,
B.K.Shoichet,
and
W.J.Tang
(2003).
Structure-based inhibitor discovery against adenylyl cyclase toxins from pathogenic bacteria that cause anthrax and whooping cough.
|
| |
J Biol Chem,
278,
25990-25997.
|
 |
|
|
|
|
 |
C.W.Dessauer,
M.Chen-Goodspeed,
and
J.Chen
(2002).
Mechanism of Galpha i-mediated inhibition of type V adenylyl cyclase.
|
| |
J Biol Chem,
277,
28823-28829.
|
 |
|
|
|
|
 |
M.A.D'Angelo,
A.E.Montagna,
S.Sanguineti,
H.N.Torres,
and
M.M.Flawiá
(2002).
A novel calcium-stimulated adenylyl cyclase from Trypanosoma cruzi, which interacts with the structural flagellar protein paraflagellar rod.
|
| |
J Biol Chem,
277,
35025-35034.
|
 |
|
|
|
|
 |
T.Onda,
Y.Hashimoto,
M.Nagai,
H.Kuramochi,
S.Saito,
H.Yamazaki,
Y.Toya,
I.Sakai,
C.J.Homcy,
K.Nishikawa,
and
Y.Ishikawa
(2001).
Type-specific regulation of adenylyl cyclase. Selective pharmacological stimulation and inhibition of adenylyl cyclase isoforms.
|
| |
J Biol Chem,
276,
47785-47793.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |
|

