 |
PDBsum entry 1ykl

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1ykl
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|

|

|

|

|

|





 (+ 0 more)
200 a.a.
(+ 0 more)
200 a.a.
|
 |

|

|

|

|

|

|

|





 (+ 0 more)
238 a.a.
(+ 0 more)
238 a.a.
|
 |

|

|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Protocatechuate 3,4-dioxygenase y408c mutant bound to dhb
|
|
Structure:
|
 |
Protocatechuate 3,4-dioxygenase alpha chain. Chain: a, c, e, g, i, k. Synonym: 3,4-pcd. Engineered: yes. Protocatechuate 3,4-dioxygenase beta chain. Chain: b, d, f, h, j, l. Synonym: 3,4-pcd. Engineered: yes. Mutation: yes
|
|
Source:
|
 |
Pseudomonas putida. Organism_taxid: 303. Gene: pcag. Expressed in: escherichia coli. Expression_system_taxid: 562. Gene: pcah. Expression_system_taxid: 562
|
|
Biol. unit:
|
 |
 24mer (from PDB file)
24mer (from PDB file)
|
|
Resolution:
|
 |
|
2.25Å
|
R-factor:
|
0.158
|
R-free:
|
0.199
|
|
|
Authors:
|
 |
C.K.Brown,D.H.Ohlendorf
|
Key ref:

|
 |
M.P.Valley
et al.
(2005).
Roles of the equatorial tyrosyl iron ligand of protocatechuate 3,4-dioxygenase in catalysis.
Biochemistry,
44,
11024-11039.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
18-Jan-05
|
Release date:
|
16-Aug-05
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chains A, B, C, D, E, F, G, H, I, J, K, L:
E.C.1.13.11.3
- protocatechuate 3,4-dioxygenase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Benzoate Metabolism
|
 |
 |
 |
 |
 |
Reaction:
|
 |
3,4-dihydroxybenzoate + O2 = 3-carboxy-cis,cis-muconate + 2 H+
|
 |
 |
 |
 |
 |
3,4-dihydroxybenzoate
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
 O2
O2
|
=
|
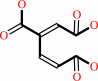 3-carboxy-cis,cis-muconate
3-carboxy-cis,cis-muconate
|
+
|
2
×
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Fe cation
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
44:11024-11039
(2005)
|
|
PubMed id:
|
|
|
|

|
| |
|
Roles of the equatorial tyrosyl iron ligand of protocatechuate 3,4-dioxygenase in catalysis.
|
|
M.P.Valley,
C.K.Brown,
D.L.Burk,
M.W.Vetting,
D.H.Ohlendorf,
J.D.Lipscomb.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The active site Fe(III) of protocatechuate 3,4-dioxygenase (3,4-PCD) from
Pseudomonas putida is ligated axially by Tyr447 and His462 and equatorially by
Tyr408, His460, and OH(-). Tyr447 and OH(-) are displaced as protocatechuate
(3,4-dihydroxybenzoate, PCA) chelates the iron and appear to serve as in situ
bases to promote this process. The role(s) of Tyr408 is (are) explored here
using mutant enzymes that exhibit less than 0.1% wild-type activity. The X-ray
crystal structures of the mutants and their PCA complexes show that the new
shorter residues in the 408 position cannot ligate the iron and instead interact
with the iron through solvents. Moreover, PCA binds as a monodentate rather than
a bidentate ligand, and Tyr447 fails to dissociate. Although the new residues at
position 408 do not directly bind to the iron, large changes in the
spectroscopic and catalytic properties are noted among the mutant enzymes.
Resonance Raman features show that the Fe-O bond of the monodentate
4-hydroxybenzoate (4HB) inhibitor complex is significantly stronger in the
mutants than in wild-type 3,4-PCD. Transient kinetic studies show that PCA and
4HB bind to 3,4-PCD in a fast, reversible step followed by a step in which
coordination to the metal occurs; the latter process is at least 50-fold slower
in the mutant enzymes. It is proposed that, in wild-type 3,4-PCD, the Lewis base
strength of Tyr408 lowers the Lewis acidity of the iron to foster the rapid
exchange of anionic ligands during the catalytic cycle. Accordingly, the
increase in Lewis acidity of the iron caused by substitution of this residue by
solvent tends to make the iron substitution inert. Tyr447 cannot be released to
allow formation of the usual dianionic PCA chelate complex with the active site
iron, and the rate of electrophilic attack by O(2) becomes rate limiting
overall. The structures of the PCA complexes of these mutant enzymes show that
hydrogen-bonding interactions between the new solvent ligand and the new
second-sphere residue in position 408 allow this residue to significantly
influence the spectroscopic and kinetic properties of the enzymes.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
N.Anitha,
and
M.Palaniandavar
(2011).
Mononuclear iron(III) complexes of 3N ligands in organized assemblies: spectral and redox properties and attainment of regioselective extradiol dioxygenase activity.
|
| |
Dalton Trans,
40,
1888-1901.
|
 |
|
|
|
|
 |
R.Mayilmurugan,
M.Sankaralingam,
E.Suresh,
and
M.Palaniandavar
(2010).
Novel square pyramidal iron(III) complexes of linear tetradentate bis(phenolate) ligands as structural and reactive models for intradiol-cleaving 3,4-PCD enzymes: Quinone formation vs. intradiol cleavage.
|
| |
Dalton Trans,
39,
9611-9625.
|
 |
|
|
|
|
 |
M.Y.Pau,
M.I.Davis,
A.M.Orville,
J.D.Lipscomb,
and
E.I.Solomon
(2007).
Spectroscopic and electronic structure study of the enzyme-substrate complex of intradiol dioxygenases: substrate activation by a high-spin ferric non-heme iron site.
|
| |
J Am Chem Soc,
129,
1944-1958.
|
 |
|
|
|
|
 |
R.F.Abdelhamid,
Y.Obara,
Y.Uchida,
T.Kohzuma,
D.M.Dooley,
D.E.Brown,
and
H.Hori
(2007).
Pi-pi interaction between aromatic ring and copper-coordinated His81 imidazole regulates the blue copper active-site structure.
|
| |
J Biol Inorg Chem,
12,
165-173.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |
|

