 |
PDBsum entry 1ux1

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase
|
 |
|
Title:
|
 |
Bacillus subtilis cytidine deaminase with a cys53his and an arg56gln substitution
|
|
Structure:
|
 |
Cytidine deaminase. Chain: a, b, c, d. Synonym: cytidine aminohydrolase, cda. Engineered: yes. Mutation: yes
|
|
Source:
|
 |
Bacillus subtilis. Organism_taxid: 1423. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Biol. unit:
|
 |
Tetramer (from PDB file)
|
|
Resolution:
|
 |
|
2.36Å
|
R-factor:
|
0.213
|
R-free:
|
0.245
|
|
|
Authors:
|
 |
E.Johansson,J.Neuhard,M.Willemoes,S.Larsen
|
Key ref:

|
 |
E.Johansson
et al.
(2004).
Structural, kinetic, and mutational studies of the zinc ion environment in tetrameric cytidine deaminase.
Biochemistry,
43,
6020-6029.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
18-Feb-04
|
Release date:
|
20-May-04
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P19079
(CDD_BACSU) -
Cytidine deaminase from Bacillus subtilis (strain 168)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
136 a.a.
131 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 2 residue positions (black
crosses)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.5.4.5
- cytidine deaminase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
cytidine + H2O + H+ = uridine + NH4+
|
|
2.
|
2'-deoxycytidine + H2O + H+ = 2'-deoxyuridine + NH4+
|
|
 |
 |
 |
 |
 |
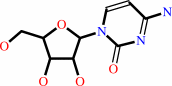 cytidine
cytidine
|
+
|
H2O
|
+
|
H(+)
|
=
|
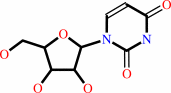 uridine
uridine
|
+
|
NH4(+)
Bound ligand (Het Group name = )
matches with 94.12% similarity
|
|
 |
 |
 |
 |
 |
 2'-deoxycytidine
2'-deoxycytidine
|
+
|
H2O
|
+
|
H(+)
|
=
|
2'-deoxyuridine
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
NH4(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Zn(2+)
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
43:6020-6029
(2004)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural, kinetic, and mutational studies of the zinc ion environment in tetrameric cytidine deaminase.
|
|
E.Johansson,
J.Neuhard,
M.Willemoës,
S.Larsen.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The zinc-containing cytidine deaminase (CDA, EC 3.5.4.5) is a pyrimidine salvage
enzyme catalyzing the hydrolytic deamination of cytidine and 2'-deoxycytidine
forming uridine and 2'-deoxyuridine, respectively. Homodimeric CDA (D-CDA) and
homotetrameric CDA (T-CDA) both contain one zinc ion per subunit coordinated to
the catalytic water molecule. The zinc ligands in D-CDA are one histidine and
two cysteine residues, whereas in T-CDA zinc is coordinated to three cysteines.
Two of the zinc coordinating cysteines in T-CDA form hydrogen bonds to the
conserved residue Arg56, and this residue together with the dipole moments from
two alpha-helices partially neutralizes the additional negative charge in the
active site, leading to a catalytic activity similar to D-CDA. Arg56 has been
substituted by a glutamine (R56Q), the corresponding residue in D-CDA, an
alanine (R56A), and an aspartate (R56D). Moreover, one of the zinc-liganding
cysteines has been substituted by histidine to mimic D-CDA, alone (C53H) and in
combination with R56Q (C53H/R56Q). R56A, R56Q, and C53H/R56Q contain the same
amount of zinc as the wild-type enzyme. The zinc-binding capacity of R56D is
reduced. Only R56A, R56Q, and C53H/R56Q yielded measurable CDA activity, R56A
and R56Q with similar K(m) but decreased V(max) values compared to wild-type
enzyme. Because of dissociation into its inactive subunits, it was impossible to
determine the kinetic parameters for C53H/R56Q. R56A and C53H/R56Q display
increased apparent pK(a) values compared to the wild-type enzyme and R56Q. On
the basis of the structures of R56A, R56Q, and C53H/R56Q an explanation is
provided of kinetic results and the apparent instability of C53H/R56Q.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
Z.A.Sánchez-Quitian,
L.F.Timmers,
R.A.Caceres,
J.G.Rehm,
C.E.Thompson,
L.A.Basso,
W.F.de Azevedo,
and
D.S.Santos
(2011).
Crystal structure determination and dynamic studies of Mycobacterium tuberculosis Cytidine deaminase in complex with products.
|
| |
Arch Biochem Biophys,
509,
108-115.
|
 |
|
|
|
|
 |
J.Norton,
H.Matsuo,
and
S.J.Sturla
(2009).
Synthesis of deoxytetrahydrouridine.
|
| |
J Org Chem,
74,
2221-2223.
|
 |
|
|
|
|
 |
S.Vincenzetti,
B.Quadrini,
P.Mariani,
G.De Sanctis,
N.Cammertoni,
V.Polzonetti,
S.Pucciarelli,
P.Natalini,
and
A.Vita
(2008).
Modulation of human cytidine deaminase by specific aminoacids involved in the intersubunit interactions.
|
| |
Proteins,
70,
144-156.
|
 |
|
|
|
|
 |
Y.Kim,
N.Maltseva,
I.Dementieva,
F.Collart,
D.Holzle,
and
A.Joachimiak
(2006).
Crystal structure of hypothetical protein YfiH from Shigella flexneri at 2 A resolution.
|
| |
Proteins,
63,
1097-1101.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

