 |
PDBsum entry 1qvv

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Structural genomics, unknown function
|
PDB id
|
|

|

|
1qvv
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Structural genomics, unknown function
|
 |
|
Title:
|
 |
Crystal structure of the s. Cerevisiae ydr533c protein
|
|
Structure:
|
 |
Ydr533c protein. Chain: a, b, c, d. Engineered: yes
|
|
Source:
|
 |
Saccharomyces cerevisiae. Baker's yeast. Organism_taxid: 4932. Gene: ydr533c. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
2.35Å
|
R-factor:
|
0.228
|
R-free:
|
0.283
|
|
|
Authors:
|
 |
M.Graille,N.Leulliot,S.Quevillon-Cheruel,H.Van Tilbeurgh
|
Key ref:

|
 |
M.Graille
et al.
(2004).
Crystal structure of the YDR533c S. cerevisiae protein, a class II member of the Hsp31 family.
Structure,
12,
839-847.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
29-Aug-03
|
Release date:
|
30-Mar-04
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q04432
(HSP31_YEAST) -
Glutathione-independent glyoxalase HSP31 from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
237 a.a.
234 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.2.1.130
- D-lactate dehydratase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
methylglyoxal + H2O = (R)-lactate + H+
|
 |
 |
 |
 |
 |
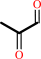 methylglyoxal
methylglyoxal
|
+
|
H2O
|
=
|
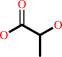 (R)-lactate
(R)-lactate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Structure
12:839-847
(2004)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of the YDR533c S. cerevisiae protein, a class II member of the Hsp31 family.
|
|
M.Graille,
S.Quevillon-Cheruel,
N.Leulliot,
C.Z.Zhou,
I.L.de la Sierra Gallay,
L.Jacquamet,
J.L.Ferrer,
D.Liger,
A.Poupon,
J.Janin,
H.van Tilbeurgh.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The ORF YDR533c from Saccharomyces cerevisiae codes for a 25.5 kDa protein of
unknown biochemical function. Transcriptome analysis of yeast has shown that
this gene is activated in response to various stress conditions together with
proteins belonging to the heat shock family. In order to clarify its biochemical
function, we determined the crystal structure of YDR533c to 1.85 A resolution by
the single anomalous diffraction method. The protein possesses an alpha/beta
hydrolase fold and a putative Cys-His-Glu catalytic triad common to a large
enzyme family containing proteases, amidotransferases, lipases, and esterases.
The protein has strong structural resemblance with the E. coli Hsp31 protein and
the intracellular protease I from Pyrococcus horikoshii, which are considered
class I and class III members of the Hsp31 family, respectively. Detailed
structural analysis strongly suggests that the YDR533c protein crystal structure
is the first one of a class II member of the Hsp31 family.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |
Figure 3.
Figure 3. The Putative Catalytic Triad(A) Hydrophobic
surface representation of the YDR533c dimer. Polar and
hydrophobic residues are colored in white and red, respectively.
The cysteine 138 side chain is shown as sticks. The putative
active site pocket from one monomer is highlighted by dotted
lines.(B) Superimposition of the YDR533c (green), E. coli Hsp31
(yellow), and PhPI (blue) catalytic triads (the proper triad
acidic residue corresponds to Glu170). The DJ-1 putative
catalytic dyad is show in pink. Only the YD533c numbering is
indicated.
|
 |
|
|

|
| |
The above figure is
reprinted
by permission from Cell Press:
Structure
(2004,
12,
839-847)
copyright 2004.
|
|
| |
Figure was
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
T.C.Terwilliger,
D.Stuart,
and
S.Yokoyama
(2009).
Lessons from structural genomics.
|
| |
Annu Rev Biophys,
38,
371-383.
|
 |
|
|
|
|
 |
F.R.Salsbury,
S.T.Knutson,
L.B.Poole,
and
J.S.Fetrow
(2008).
Functional site profiling and electrostatic analysis of cysteines modifiable to cysteine sulfenic acid.
|
| |
Protein Sci,
17,
299-312.
|
 |
|
|
|
|
 |
K.Goyal,
and
S.C.Mande
(2008).
Exploiting 3D structural templates for detection of metal-binding sites in protein structures.
|
| |
Proteins,
70,
1206-1218.
|
 |
|
|
|
|
 |
W.Liu,
Y.Y.Zhou,
M.K.Teng,
and
C.Z.Zhou
(2007).
Purification, crystallization and preliminary X-ray analysis of Hsp33 from Saccharomyces cerevisiae.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
63,
114-116.
|
 |
|
|
|
|
 |
S.Quevillon-Cheruel,
N.Leulliot,
M.Graille,
K.Blondeau,
J.Janin,
and
H.van Tilbeurgh
(2006).
Crystal structure of the yeast His6 enzyme suggests a reaction mechanism.
|
| |
Protein Sci,
15,
1516-1521.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
T.Miura,
H.Minegishi,
R.Usami,
and
F.Abe
(2006).
Systematic analysis of HSP gene expression and effects on cell growth and survival at high hydrostatic pressure in Saccharomyces cerevisiae.
|
| |
Extremophiles,
10,
279-284.
|
 |
|
|
|
|
 |
C.Z.Zhou,
P.Meyer,
S.Quevillon-Cheruel,
I.L.De La Sierra-Gallay,
B.Collinet,
M.Graille,
K.Blondeau,
J.M.François,
N.Leulliot,
I.Sorel,
A.Poupon,
J.Janin,
and
H.Van Tilbeurgh
(2005).
Crystal structure of the YML079w protein from Saccharomyces cerevisiae reveals a new sequence family of the jelly-roll fold.
|
| |
Protein Sci,
14,
209-215.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Quevillon-Cheruel,
N.Leulliot,
M.Graille,
N.Hervouet,
F.Coste,
H.Bénédetti,
C.Zelwer,
J.Janin,
and
H.Van Tilbeurgh
(2005).
Crystal structure of yeast YHR049W/FSH1, a member of the serine hydrolase family.
|
| |
Protein Sci,
14,
1350-1356.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

