 |
PDBsum entry 1nhc

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase
|
 |
|
Title:
|
 |
Structural insights into the processivity of endopolygalacturonase i from aspergillus niger
|
|
Structure:
|
 |
Polygalacturonase i. Chain: a, b, c, d, e, f. Synonym: pg-i, pectinase, endopolygalacturonase i. Engineered: yes
|
|
Source:
|
 |
Aspergillus niger. Organism_taxid: 5061. Expressed in: aspergillus niger. Expression_system_taxid: 5061
|
|
Resolution:
|
 |
|
1.70Å
|
R-factor:
|
0.176
|
R-free:
|
0.209
|
|
|
Authors:
|
 |
G.Van Pouderoyen,H.J.Snijder,J.A.Benen,B.W.Dijkstra
|
Key ref:

|
 |
G.van Pouderoyen
et al.
(2003).
Structural insights into the processivity of endopolygalacturonase I from Aspergillus niger.
FEBS Lett,
554,
462-466.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
19-Dec-02
|
Release date:
|
25-Nov-03
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P26213
(PGLR1_ASPNG) -
Endopolygalacturonase I from Aspergillus niger
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
368 a.a.
336 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.2.1.15
- endo-polygalacturonase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(1,4-alpha-D-galacturonosyl)n+m + H2O = (1,4-alpha-D-galacturonosyl)n + (1,4-alpha-D-galacturonosyl)m
|
 |
 |
 |
 |
 |
(1,4-alpha-D-galacturonosyl)n+m
|
+
|
H2O
|
=
|
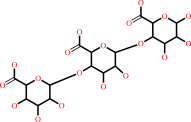 (1,4-alpha-D-galacturonosyl)n
(1,4-alpha-D-galacturonosyl)n
|
+
|
(1,4-alpha-D-galacturonosyl)m
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
FEBS Lett
554:462-466
(2003)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural insights into the processivity of endopolygalacturonase I from Aspergillus niger.
|
|
G.van Pouderoyen,
H.J.Snijder,
J.A.Benen,
B.W.Dijkstra.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Endopolygalacturonase I is a processive enzyme, while the 60% sequence identical
endopolygalacturonase II is not. The 1.70 A resolution crystal structure of
endopolygalacturonase I reveals a narrowed substrate binding cleft. In addition,
Arg96, a residue in this cleft previously shown to be critical for processivity,
interacts with the substrate mimics glycerol and sulfate in several well-defined
conformations in the six molecules in the asymmetric unit. From this we conclude
that both Arg96 and the narrowed substrate binding cleft contribute to retaining
the substrate while it moves through the active site after a cleavage event has
occurred.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Fig. 1. The three-dimensional structure of (A)
endopolygalacturonase I and (B) endopolygalacturonase II with
the N- and C-termini indicated, viewed onto β-sheet PB2a
(yellow). PB2b (blue) is the bottom β-sheet. PB1 is shown in
green and PB3 is shown in red. The active site (Asp186, Asp207
and Asp208) is visible at the top, between the T1 loop regions
(on the right side) and the T3 loop regions (on the left side).
The glycosylation of endopolygalacturonase I and the loops
bordering the active site cleft are shown in ball-and-stick
representation (endopolygalacturonase I residues 124–128
(left) and 299–301 (right), endopolygalacturonase II residues
121–123 (left) and 293–295 (right)). The smallest distance
at the entry of the active site cleft is indicated with a dashed
line and the value is shown [34]. (For interpretation of the
references to color in this figure legend, the reader is
referred to the web version of this article.)
|
 |
Figure 2.
Fig. 2. N-Glycosylation at Asn246.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the Federation of European Biochemical Societies:
FEBS Lett
(2003,
554,
462-466)
copyright 2003.
|
|
| |
Figures were
selected
by the author.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
D.D.Sprockett,
H.Piontkivska,
and
C.B.Blackwood
(2011).
Evolutionary analysis of glycosyl hydrolase family 28 (GH28) suggests lineage-specific expansions in necrotrophic fungal pathogens.
|
| |
Gene,
479,
29-36.
|
 |
|
|
|
|
 |
W.X.Sun,
Y.J.Jia,
B.Z.Feng,
N.R.O'Neill,
X.P.Zhu,
B.Y.Xie,
and
X.G.Zhang
(2009).
Functional analysis of Pcipg2 from the straminopilous plant pathogen Phytophthora capsici.
|
| |
Genesis,
47,
535-544.
|
 |
|
|
|
|
 |
D.W.Abbott,
and
A.B.Boraston
(2008).
Structural biology of pectin degradation by Enterobacteriaceae.
|
| |
Microbiol Mol Biol Rev,
72,
301.
|
 |
|
|
|
|
 |
H.J.Rozeboom,
T.M.Bjerkan,
K.H.Kalk,
H.Ertesvåg,
S.Holtan,
F.L.Aachmann,
S.Valla,
and
B.W.Dijkstra
(2008).
Structural and Mutational Characterization of the Catalytic A-module of the Mannuronan C-5-epimerase AlgE4 from Azotobacter vinelandii.
|
| |
J Biol Chem,
283,
23819-23828.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
H.Trigui-Lahiani,
M.Ayadi,
N.Hadj-Taïeb,
M.B.Ali,
and
A.Gargouri
(2008).
Genomic organization of a polygalacturonase gene from a hyperpectinolytic mutant strain of Penicillium occitanis.
|
| |
FEMS Microbiol Lett,
281,
23-29.
|
 |
|
|
|
|
 |
M.do Rosário Freixo,
A.Karmali,
and
J.M.Arteiro
(2008).
Production of polygalacturonase from Coriolus versicolor grown on tomato pomace and its chromatographic behaviour on immobilized metal chelates.
|
| |
J Ind Microbiol Biotechnol,
35,
475-484.
|
 |
|
|
|
|
 |
P.B.Vordtriede,
and
M.D.Yoder
(2008).
Crystallization, X-ray diffraction analysis and preliminary structure determination of the polygalacturonase PehA from Agrobacterium vitis.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
64,
645-647.
|
 |
|
|
|
|
 |
L.Federici,
A.Di Matteo,
J.Fernandez-Recio,
D.Tsernoglou,
and
F.Cervone
(2006).
Polygalacturonase inhibiting proteins: players in plant innate immunity?
|
| |
Trends Plant Sci,
11,
65-70.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

