 |
PDBsum entry 1mr2

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase
|
 |
|
Title:
|
 |
Structure of the mt-adprase in complex with 1 mn2+ ion and amp-cp (a inhibitor), a nudix enzyme
|
|
Structure:
|
 |
Adpr pyrophosphatase. Chain: a. Synonym: adprase, mutt/nudix family protein, hypothetical protein rv1700. Engineered: yes
|
|
Source:
|
 |
Mycobacterium tuberculosis. Organism_taxid: 1773. Gene: rv1700. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Biol. unit:
|
 |
 Dimer (from PDB file)
Dimer (from PDB file)
|
|
Resolution:
|
 |
|
2.30Å
|
R-factor:
|
0.205
|
R-free:
|
0.260
|
|
|
Authors:
|
 |
L.-W.Kang,S.B.Gabelli,M.A.Bianchet,J.E.Cunningham,S.F.O'Handley, L.M.Amzel
|
Key ref:

|
 |
L.W.Kang
et al.
(2003).
Structure and mechanism of MT-ADPRase, a nudix hydrolase from Mycobacterium tuberculosis.
Structure,
11,
1015-1023.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
17-Sep-02
|
Release date:
|
05-Aug-03
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
O33199
(O33199_MYCTO) -
MutT/nudix family protein from Mycobacterium tuberculosis (strain CDC 1551 / Oshkosh)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
207 a.a.
187 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.6.1.13
- ADP-ribose diphosphatase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
ADP-D-ribose + H2O = D-ribose 5-phosphate + AMP + 2 H+
|
 |
 |
 |
 |
 |
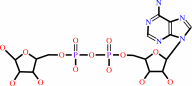 ADP-D-ribose
ADP-D-ribose
|
+
|
H2O
|
=
|
D-ribose 5-phosphate
Bound ligand (Het Group name = )
matches with 78.57% similarity
|
+
|
 AMP
AMP
|
+
|
2
×
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Structure
11:1015-1023
(2003)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structure and mechanism of MT-ADPRase, a nudix hydrolase from Mycobacterium tuberculosis.
|
|
L.W.Kang,
S.B.Gabelli,
J.E.Cunningham,
S.F.O'Handley,
L.M.Amzel.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Nudix hydrolases are a family of proteins that contain the characteristic
sequence GX(5)EX(7)REUXEEXG(I/L/V), the Nudix box. They catalyze the hydrolysis
of a variety of nucleoside diphosphate derivatives such as ADP-ribose, Ap(n)A (3
</= n </= 6), NADH, and dATP. A number of Nudix hydrolases from several
species, ranging from bacteria to humans, have been characterized, including, in
some cases, the determination of their three-dimensional structures. The product
of the Rv1700 gene of M. tuberculosis is a Nudix hydrolase specific for
ADP-ribose (ADPR). We have determined the crystal structures of MT-ADPRase
alone, and in complex with substrate, with substrate and the nonactivating metal
ion Gd(3+), and in complex with a nonhydrolyzable ADPR analog and the activating
metal ion Mn(2+). These structures, refined with data extending to resolutions
between 2.0 and 2.3 A, showed that there are sequence differences in binding
site residues between MT-ADPRase and a human homolog that may be exploited for
antituberculosis drug development.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |
Figure 5.
Figure 5. Recognition of the Nonhydrolyzable Substrate
Analog, AMPCPR, and Three Mn2+ Ions by MT-ADPRase(A) Schematic
representation. Residue 142 is from loop L9 that is not ordered
without three manganese ions and AMPCPR.(B) Stereo diagram of
the ADP-ribose binding site with AMPCPR and three manganese
ions. Residues and substrate are shown using the same
representation and color scheme of Figure 4A. Broken orange
lines represent polar interactions among AMPCPR, three manganese
ions, and protein residues.
|
 |
|
|

|
| |
The above figure is
reprinted
by permission from Cell Press:
Structure
(2003,
11,
1015-1023)
copyright 2003.
|
|
| |
Figure was
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
T.Nakamura,
S.Meshitsuka,
S.Kitagawa,
N.Abe,
J.Yamada,
T.Ishino,
H.Nakano,
T.Tsuzuki,
T.Doi,
Y.Kobayashi,
S.Fujii,
M.Sekiguchi,
and
Y.Yamagata
(2010).
Structural and dynamic features of the MutT protein in the recognition of nucleotides with the mutagenic 8-oxoguanine base.
|
| |
J Biol Chem,
285,
444-452.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.G.Thorsell,
C.Persson,
S.Gräslund,
M.Hammarström,
R.D.Busam,
and
B.M.Hallberg
(2009).
Crystal structure of human diphosphoinositol phosphatase 1.
|
| |
Proteins,
77,
242-246.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
N.Huang,
J.De Ingeniis,
L.Galeazzi,
C.Mancini,
Y.D.Korostelev,
A.B.Rakhmaninova,
M.S.Gelfand,
D.A.Rodionov,
N.Raffaelli,
and
H.Zhang
(2009).
Structure and function of an ADP-ribose-dependent transcriptional regulator of NAD metabolism.
|
| |
Structure,
17,
939-951.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.A.Messing,
S.B.Gabelli,
Q.Liu,
H.Celesnik,
J.G.Belasco,
S.A.Piñeiro,
and
L.M.Amzel
(2009).
Structure and biological function of the RNA pyrophosphohydrolase BdRppH from Bdellovibrio bacteriovorus.
|
| |
Structure,
17,
472-481.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
G.W.Buchko,
O.Litvinova,
H.Robinson,
A.F.Yakunin,
and
M.A.Kennedy
(2008).
Functional and structural characterization of DR_0079 from Deinococcus radiodurans, a novel Nudix hydrolase with a preference for cytosine (deoxy)ribonucleoside 5'-Di- and triphosphates.
|
| |
Biochemistry,
47,
6571-6582.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
N.Huang,
L.Sorci,
X.Zhang,
C.A.Brautigam,
X.Li,
N.Raffaelli,
G.Magni,
N.V.Grishin,
A.L.Osterman,
and
H.Zhang
(2008).
Bifunctional NMN adenylyltransferase/ADP-ribose pyrophosphatase: structure and function in bacterial NAD metabolism.
|
| |
Structure,
16,
196-209.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
T.Wakamatsu,
N.Nakagawa,
S.Kuramitsu,
and
R.Masui
(2008).
Structural basis for different substrate specificities of two ADP-ribose pyrophosphatases from Thermus thermophilus HB8.
|
| |
J Bacteriol,
190,
1108-1117.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.Jung,
Y.J.Ahn,
and
L.W.Kang
(2007).
Overexpression, crystallization and preliminary X-ray crystallographic analysis of Nudix hydrolase Orf141 from Escherichia coli K-1.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
63,
812-815.
|
 |
|
|
|
|
 |
S.B.Gabelli,
M.A.Bianchet,
W.Xu,
C.A.Dunn,
Z.D.Niu,
L.M.Amzel,
and
M.J.Bessman
(2007).
Structure and function of the E. coli dihydroneopterin triphosphate pyrophosphatase: a Nudix enzyme involved in folate biosynthesis.
|
| |
Structure,
15,
1014-1022.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.K.Arakaki,
W.Tian,
and
J.Skolnick
(2006).
High precision multi-genome scale reannotation of enzyme function by EFICAz.
|
| |
BMC Genomics,
7,
315.
|
 |
|
|
|
|
 |
D.I.Fisher,
J.L.Cartwright,
and
A.G.McLennan
(2006).
Characterization of the Mn2+-stimulated (di)adenosine polyphosphate hydrolase encoded by the Deinococcus radiodurans DR2356 nudix gene.
|
| |
Arch Microbiol,
186,
415-424.
|
 |
|
|
|
|
 |
K.Okuda,
H.Hayashi,
and
Y.Nishiyama
(2005).
Systematic characterization of the ADP-ribose pyrophosphatase family in the Cyanobacterium Synechocystis sp. strain PCC 6803.
|
| |
J Bacteriol,
187,
4984-4991.
|
 |
|
|
|
|
 |
G.W.Buchko,
S.Ni,
S.R.Holbrook,
and
M.A.Kennedy
(2004).
Solution structure of hypothetical Nudix hydrolase DR0079 from extremely radiation-resistant Deinococcus radiodurans bacterium.
|
| |
Proteins,
56,
28-39.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Mishima,
Y.Sakai,
N.Itoh,
H.Kamiya,
M.Furuichi,
M.Takahashi,
Y.Yamagata,
S.Iwai,
Y.Nakabeppu,
and
M.Shirakawa
(2004).
Structure of human MTH1, a Nudix family hydrolase that selectively degrades oxidized purine nucleoside triphosphates.
|
| |
J Biol Chem,
279,
33806-33815.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.B.Gabelli,
M.A.Bianchet,
H.F.Azurmendi,
Z.Xia,
V.Sarawat,
A.S.Mildvan,
and
L.M.Amzel
(2004).
Structure and mechanism of GDP-mannose glycosyl hydrolase, a Nudix enzyme that cleaves at carbon instead of phosphorus.
|
| |
Structure,
12,
927-935.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.Yoshiba,
T.Ooga,
N.Nakagawa,
T.Shibata,
Y.Inoue,
S.Yokoyama,
S.Kuramitsu,
and
R.Masui
(2004).
Structural insights into the Thermus thermophilus ADP-ribose pyrophosphatase mechanism via crystal structures with the bound substrate and metal.
|
| |
J Biol Chem,
279,
37163-37174.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

